Abstract
This study aimed to determine the total phenolic content, DPPH scavenging, α-glucosidase, and nitric oxide (NO) inhibition of Gynura procumbens and Cleome gynandra extracts obtained with five different ethanolic concentrations. The findings showed that the 100% ethanolic extract of G. procumbens had the highest phenolic content and the lowest IC50 values for DPPH scavenging and NO inhibition activity compared to the properties of the other extracts. For C. gynandra, the 20% and 100% ethanolic extracts had comparably high total phenolic contents, and the latter possessed the lowest IC50 value in the NO inhibition assay. In addition, the 20% ethanolic extract of C. gynandra had the lowest IC50 value in the DPPH scavenging assay. However, none of the extracts from either herb had the ability to inhibit α-glucosidase enzyme. Pearson correlation analysis indicated a strong relationship between the phenolic content and DPPH scavenging activity in both herb extracts. A moderately strong relationship was also observed between the phenolic content and NO inhibition in G. procumbens extracts and not in C. gynandra extracts. The UHPLC-ESI-Orbitrap-MS revealed major phenolics from the groups of hydroxycinnamic acids, hydroxybenzoic acids, and flavonoid derivatives from both herbs, which could be the key contributors to their bioactivities. Among the identified metabolites, 24 metabolites were tentatively assigned for the first time from both species of studied herbs. These two herbs could be recommended as prospective natural products with valuable medicinal properties.
1. Introduction
Plants and herbs have a history of traditional uses and are important parts of cultural heritage. Their appreciation as food and links to health-promoting benefits are also significant. Since ancient times, herbs have been utilised traditionally to cure many illnesses, which has prompted modern science to fully understand their benefits. Gynura procumbens (Asteraceae), locally known as Sambung nyawa, is an annual grown herb that has thick leaves and hardened stems with a slight purple tint during maturation. The young leaves can be eaten raw as salads. Its ethnomedicinal usages are well reported; for example, in Indonesia, G. procumbens is used to treat fever, skin rashes, and ringworm infection [1]. In Thailand, it is used to treat inflammation, viral infections, and rheumatism [2]. Scientific investigations on G. procumbens include antidiabetic, antihypertensive [3], anticancer [4], and anti-inflammatory [5] studies. In terms of its phytochemical constituents, phenolic compounds, such as kaempferol, quercetin, astragalin [6], kaempferol 3-O-rutinoside, rutin, and chlorogenic acid [7], were identified as the key metabolites that contributed to the bioactivities in the reported studies.
Cleome gynandra (Cleomaceae), known as maman, is used in ayurvedic treatment, especially for treating lump, prostate enlargement, worm infections, and ear diseases [8]. The leaves of C. gynandra are crushed into a concoction and drunk to cure scurvy in Africa [8]. Scientific investigations on the biological activities of C. gynandra include antioxidant [9], anti-inflammatory [10], antidiabetic [11], and anticancer [12]. However, phytochemical analysis is still lacking for C. gynandra, except in a recent report that listed caffeic acid, coumaric acid, sinapic acid, and ferulic acid as the major phytochemicals in this herb [13]. Ranjitha and colleagues [14] identified β-amyrin, β-amyrin-3-O-β-glucopyranoside, sitosterol, and stigmasterol by NMR and GC-MS from this herb. The C. gynandra was reported to be rich in minerals, carotenoids, flavonoids, alkaloids, terpenoids, steroids, and tannins.
Inflammation is a coordinated response in the body towards harmful stimuli, such as injuries, pathogenic infection, and allergens. Macrophages are the cells responsible for initiating inflammation by producing inflammatory mediators, such as cytokines, interferons, and nitric oxide (NO) as response to stress [15]. Study has also indicated how the production of excessive inflammatory mediators leads to the onset of diabetes [16]. Among the treatments available for diabetes, the inhibition of α-glucosidase is one of them, which is responsible for glucose breakdown in the intestinal wall [17]. Plants' phenolic compounds have been reported to have the ability in inhibiting this enzyme, yet more often, modern drugs can be prescribed to combat the occurrence of inflammation-related diseases and diabetes [18]. However, prolonged use of these drugs stimulates unwanted side effects towards the liver, kidney, and other organs [19]. In return, naturally occurring phytochemicals from herbs have been ventured as an alternative medicine since they possess reduced or no toxicity when consumed at lower doses. G. procumbens and C. gynandra are two herbs that have the potential to be explored further for their anti-inflammatory and antidiabetic properties based on past studies. An optimum and proper extraction protocol may help researchers study the beneficial health properties of herbs, thus enabling the development of herbal-based products [20]. Extraction protocols that vary based on the type of sample, extraction solvents, temperature, extraction time, and instruments used play important roles in the standardization of herbs [21].
In this study, the efficacy of G. procumbens and C. gynandra extracted with different ethanol concentrations was tested for α-glucosidase and NO inhibition. Nevertheless, detailed metabolite profile and the effect of solvents extractions on distribution of metabolites of both herbs are still lacking. As such, this study proposed to investigate the aforementioned properties of the herbal extracts. The total phenolic content (TPC) and 2,2-diphenyl-1-picrylhydrazyl (DPPH) free radical scavenging activity were also tested as to support the anti-inflammatory and antidiabetic properties of the studied herbs. The relationship between TPC and biological activity was studied using the Pearson correlation model. Active ethanolic extracts from both herbs were then subjected to ultra high-performance liquid chromatography-electron spray ionisation-orbitrap-mass spectrometry (UHPLC-ESI-Orbitrap-MS), and potential metabolites that may contribute to the tested bioactivities were tentatively identified and reported.
2. Materials and Methods
2.1. Chemicals and Reagents
HPLC-grade ethanol and Folin–Ciocalteu's reagent were purchased from Merck (Darmstadt, Germany). Sodium bicarbonate and dimethyl sulfoxide (DMSO) were supplied by Fisher Scientific (Leicestershire, UK). 2,2-Diphenyl-1-picrylhydrazyl (DPPH), quercetin hydrate, curcumin, gallic acid, 4-nitrophenyl α-D-glucopyranoside (PNPG), lipopolysaccharide (LPS), phosphate-buffered saline (PBS), and recombinant murine interferon gamma (IFN-γ) were obtained from Sigma-Aldrich (St. Louis, USA). The α-glucosidase enzyme was purchased from Megazyme (Bray Business Park, Ireland). The reagents used for cell culture studies, including Dulbecco's Modified Eagle's Medium (DMEM), containing HEPES and L-glutamine, with and without phenol red; an antibiotic mixture of penicillin-streptomycin, fetal bovine serum (FBS), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and TripLE™ Express enzyme were supplied by Gibco (Life technologies, USA). The LC/MS graded acetonitrile was purchased from Fisher Scientific (Toronto, Canada). RAW 264.7 murine macrophage cells were obtained from American Type Culture Collection (ATCC, Rockville, MD).
2.2. Plant Material
C. gynandra was collected from a local farmer in Rompin, Negeri Sembilan. Samples were randomly plucked from plots in an open area that had been preinstalled with automated irrigation. G. procumbens was grown and harvested from the net house located in the Malaysia Agricultural, Research and Development Institute (MARDI). Both herbs were collected before noon to maintain their freshness. Upon collection, voucher sample from both herbs were sent to MARDI's herbarium and authenticated by a botanist, Dr. Mohd Norfaizal Ghazalli (voucher specimen numbers for G. procumbens and C. gynandra are MDI 12841 and MDI 12840, respectively). Only the leaves from both herbs were used in this study. The leaves were washed under running tap water and gently dried using laboratory tissue paper. After that, they were ground with liquid nitrogen using a mortar and pestle and freeze-dried immediately. The dried powder samples were stored at -20 °C prior to analysis.
2.3. Extraction of Samples
Six replicates of freeze-dried samples of both G. procumbens and C. gynandra were extracted with different concentrations of ethanol/water ratios (0, 20, 50, 70, and 100% ethanol). In general, the dried samples (4 g) were mixed in 250 mL conical flasks with 100 mL of each ethanol ratio. The mixture was then homogenised using a homogeniser (Ultra Turrax, IKA, Germany) at 6000 rpm for 1 min followed by shaking in a shaker (ES-20, Biosan, Latvia) at 220 rpm for 15 min at room temperature. The supernatant was filtered with125 mm diameter filter paper (Advantec, Japan). The extraction procedures were repeated twice. The collected supernatant was concentrated using a rotary evaporator (RII, Buchi, Switzerland) and further freeze-dried (Freezone 6, Labconco, USA) to eliminate any moisture. Prior to bioassay analysis, the crude extracts of 0 and 20% ethanol/water were dissolved in deionised water, whereas the others were dissolved in dimethyl sulfoxide (DMSO).
2.4. Total Phenolic Content (TPC) Assay
The TPC assay was carried out in accordance with the method described in [22] with minor modifications. Modifications were made in terms of the volume of reagents used in the assay. In general, 20 μL of 350 μg/mL of the extract was mixed with 100 μL of Folin–Ciocalteu's reagent (10-fold dilution) in a 96-well plate. Then, 80 μL of 7.5% sodium carbonate was added, and the mixture was left in the dark for 30 min prior to the absorbance reading at 750 nm (SpectraMax PLUS, USA). Gallic acid was serially diluted ranging from 0.78 to 100 μg/mL and used to make a standard curve in this assay. The results are expressed as mg gallic acid equivalent (GAE) per 100 mg of dried extract.
2.5. 2,2-Diphenyl-1-picrylhydrazyl (DPPH) Scavenging Assay
The DPPH scavenging assay was performed based on the method described in [22] with minor modifications to the concentration and volume of DPPH used. In general, 50 μL of extract with serial dilutions ranging from 10.94 to 350 μg/mL was mixed with 100 μL of DPPH (0.15 mM) in a 96-well plate and incubated in the dark for 30 min. The absorbance of the solution was measured at 515 nm. Quercetin was used as the positive control, and all experiments were performed in six replicates. The results are expressed as the concentration (μg/mL) of extract needed to scavenge 50% of DPPH (IC50).
2.6. α-Glucosidase Inhibition Assay
The inhibition of the α-glucosidase enzyme by G. procumbens and C. gynandra extracts was evaluated based on the method described in [23] with minor modifications to the used volume of the enzyme and substrate. Prior to the experiment, α-glucosidase and the substrate PNPG were dissolved in 50 mM phosphate buffer at pH 6.5. In brief, 10 μL of plant extract with a serial dilution ranging from 10.94 to 350 μg/mL was mixed with 130 μL of 30 mM phosphate buffer and 10 μL of the enzyme (2 U/mL) in a 96-well plate. After 5 min of incubation, 50 μL of the PNPG substrate was added followed by and reincubation at room temperature for another 15 min. The reaction was ceased by the addition of 50 μL of glycine (pH 10) before the absorbance was measured at 405 nm. Quercetin was used as the positive control in this study. The results are expressed as the concentration (μg/mL) of extract needed to inhibit 50% of α-glucosidase (IC50).
2.7. Nitric Oxide (NO) Inhibition Activity
The NO inhibition assay was performed in accordance with the method described in [24]. The RAW 264.7 cells were grown in culture flasks using phenol-red DMEM under 5% CO2 at 37°C. Once confluency reached 80%, cells were detached using 2.5 mL TrypLE™ Express enzyme. Prior to cells seeding in 96-well plates, cells were counted using the standard Trypan blue counting technique, where the cell concentration was set to 1 × 104 cells/mL in all wells. Seeded cells (50 μL/well) were left in an incubator for 24 h before proceeding with induction and treatment. In all, 50 μL (1 μL IFN-γ + 1μL LPS + 48 μL DMEM) triggering agent was added into the designated wells, followed by 50 μL of plant extract (serially diluted from 15.63 to 500 μg/mL). Curcumin was used as the positive control in this assay. All analyses were performed in six replicates, and the cells were incubated 17–24 h at 37°C under a 5% CO2 atmosphere.
2.8. Measurement of Nitrite
A Griess assay was performed to measure the accumulation of nitrite ions (NO2−), a conversion product from NO in a simple manner. An incubated 96-well plate was removed, and 50 μL of media from the plate was transferred attentively into a new 96-well plate. Then, 50 μL of Griess reagent (1% sulfanilamide, 0.1% N-(1-naphtyl)-ethylene diamine dihydrochloride, and 2.5% phosphoric acid) was added, and the plate was left in the dark for 15 min at room temperature. Sodium nitrite (NaNO2) at 200 μM was used as a positive control in this assay. Absorbance at 550 nm was measured after incubation.
2.9. Cell Viability Test
Cell viability was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay to determine the cytotoxicity of the plant extract. Fresh phenol-red DMEM (100 μL) was added to the wells containing cells, followed by 20 μL of MTT (dissolved in 1× PBS buffer). The plate was then incubated for 4 h at 37°C under a 5% CO2 atmosphere. Next, all the media were discarded, and 100 μL of DMSO was added. The plate was left for 15 min in the dark at room temperature before the absorbance was measured at 570 nm. The percent viability of the cells was calculated by comparing the absorbance of the treated cells with the control group (untreated cells).
2.10. Metabolite Profiling Using the UHPLC-ESI-Orbitrap-MS
The separation and identification of metabolites from G. procumbens and C. gynandra were achieved using a UHPLC system (Ultimate 3000™, Thermo Scientific) coupled with mass spectrometry (Q Exactive™ Hybrid Quadrupole-Orbitrap, Thermo Scientific). Separation of the metabolites was performed using a Thermo C18 column (2.1 mm × 100 mm, 1.9 μm) with mobile phases of LCMS-grade acetonitrile with 0.1% formic acid as buffer B and deionised water with 0.1% formic acid as buffer A. The flow rate was set at 274 μL/min, and UV detection was set at 280 nm. The gradient setting of the mobile phases was set for a total run time of 30 min and divided as follows: equilibration of column for 5 min at 95% of solvent A, a steady decrease in solvent A for 20 min until reaching 5%, maintenance of 5 % solvent A for another 5 min, and a steep increase in solvent A to 95% in one minute. The column was re-equilibrated for 4 min at 95% solvent A. All samples were prepared at a concentration of 2.0 mg/mL in 30% methanol and filtered by a nylon syringe filter (0.45 μM, 13 mm diameter). Ten microliters of each sample was injected into the system. Ionization of the metabolites was performed using an ESI probe in negative mode. The capillary temperature was set at 300°C with a scanning range from 50–1500 amu. All analyses were performed and monitored using Xcalibur 2.2 software (Thermo Scientific Inc, Waltham, MA, USA). The identification and characterization of metabolites were performed by relative comparison from previously reported data and from online databases [25, 26]. The mass error in ppm was calculated by comparing the theoretical monoisotopic mass from the online databases to the observed mass.
2.11. Statistical Analysis
All bioassay results were reported as the mean of six biological replicates with the standard deviation. One-way analysis of variance (ANOVA) was performed with a significant difference between the collected data set at a confidence interval of 95%. The Pearson correlation test (r value) was calculated to evaluate the relationship between the bioactivities. All calculations were performed using GraphPad PRISM version 5.01 for Windows (San Diego, CA, USA).
3. Results and Discussion
3.1. Total Phenolic Content of G. procumbens and C. gynandra Extracts
The TPC results of the G. procumbens and C. gynandra extracts are presented in Table 1. The TPC assay was performed to measure the relative amounts of phenolic compounds from the herbal extracts since most phenolic compounds were found to possess health benefits [27]. G. procumbens extracted with 100% ethanol was observed to have the highest phenolic content with 5.91 mg GAE/100 mg dried extract (de). The TPC trend was 100% > 70% > 50% > 20% > 0% ethanolic extract as the ratio of water increased. On the other hand, the TPC of C. gynandra extracts showed mixed results, with 0%, 20%, and 100% ethanolic extracts showing significantly high phenolic content with 3.38, 3.48, and 3.71 mg GAE/100 mg de, respectively. The TPC values increased as the ratio of ethanol increased and started to drop in the 50% and 70% ethanolic extracts before rising again at the 100% ethanolic concentration. The difference in the TPC trend shown by the two herbs could be due to the type of metabolites being extracted. Previous study has indicated how polar metabolites in nature have the tendency to be extracted with polar solvents and vice versa [28]. Secondary metabolites, such as tannins, hydroxycinnamic acids, and flavonoids are regarded as polar in nature, whereas sterols, terpenoids, and lipids are semipolar and nonpolar [29]. Water, being a universal solvent, is also more polar than ethanol. Increasing the water ratio in ethanol makes the final concentration of the extraction solvent more polar. In this study, different water/ethanol concentrations were thought to extract metabolites of different polarities, thus giving different clusters of metabolites in each extraction solvent.
Table 1.
The total phenolic content (TPC), DPPH scavenging activity, nitric oxide (NO) inhibition activity, and cell cytotoxicity (MTT) of the G. procumbens and G. gynandra extracts.
| Extracts % EtOH | G. procumbens | C. gynandra | ||||||
|---|---|---|---|---|---|---|---|---|
| TPC (mg GAE/100 mgde) | DPPH (IC50) μg/mL | NO inhibition (IC50) μg/mL | MTT (%) at 1000 μg/mL | TPC (mg GAE/100 mgde) | DPPH (IC50) μg/mL | NO inhibition (IC50) μg/mL | MTT (%) at 1000 μg/mL | |
| 0 | 1.44 ± 0.03a | 124.37 ± 6.28a | 117.19 ± 10.50a | 96.31 ± 3.06 | 3.38 ± 0.06a | 62.01 ± 5.84a | 210.06 ± 12.25a | ND |
| 20 | 1.58 ± 0.32a | 317.41 ± 8.32b | 96.77 ± 7.09b | 97.91 ± 3.84 | 3.46 ± 0.26a | 40.36 ± 5.09b | 94.83 ± 10.49b | ND |
| 50 | 3.81 ± 0.04b | 66.28 ± 3.95c | 82.03 ± 8.95c | 95.76 ± 8.01 | 1.51 ± 0.18b | 276.92 ± 6.42c | 97.11 ± 9.09b | ND |
| 70 | 3.15 ± 0.19b | 131.92 ± 8.99a | 22.91 ± 6.75d | 93.54 ± 8.73 | 2.74 ± 0.28c | 304.64 ± 9.02d | 66.01 ± 7.75c | 91.53 ± 3.25 |
| 100 | 5.91 ± 0.62c | 63.73 ± 2.90c | 25.20 ± 6.19d | 91.71 ± 7.72 | 3.71 ± 0.26a | 236.95 ± 7.96c | 60.75 ± 6.32c | 89.11 ± 4.77 |
| Quercetin | — | 1.73 ± 0.12 | — | — | — | 1.73 ± 0.12 | — | — |
| Curcumin | — | — | 3.59 ± 0.28 | — | — | — | 3.59 ± 0.28 | — |
Values are the mean ± standard deviation of six biological replicates. Bold numbers to show extracts with the best activity for related assays. GAE: gallic acid equivalent; de: dried extract; EtOH: ethanol; ND: not detected. Each different superscript letter within the same assay indicates a statistically significant different (P < 0.05).
3.2. DPPH Scavenging Activity of the G. procumbens and C. gynandra Extracts
The ability of the ethanolic extracts of G. procumbens and C. gynandra to scavenge DPPH free radicals was tested, and the results are presented in Table 1. Unlike TPC, DPPH scavenging in the G. procumbens extracts showed fluctuating results. The 20% ethanolic extract of C. gynandra had the lowest IC50 value of 40.36 μg/mL, followed by the water extract with IC50 value of 62.01 μg/mL. The 50 and 100% ethanolic extracts of G. procumbens had the IC50 values of 66.28 μg/mL and 63.73 μg/mL, respectively, with no significant difference. The IC50 of the other ethanolic extracts were fluctuated showing the trend of 0% > 70% > 20%. In addition, the rest of the ethanolic extracts possessed poor DPPH scavenging activity, as shown in Table 1.
3.3. α-Glucosidase Inhibition Activity of the G. procumbens and C. gynandra Extracts
Unlike the rest of the assays, neither extract (G. procumbens or C. gynandra) showed any inhibition against the α-glucosidase enzyme. In this study, extracts from both plants at 10 mg/mL were tested against α-glucosidase, and no activity was detected except for the positive control, quercetin. Quercetin showed 92.3% inhibition at 50 μg/mL. This finding contradicts findings from other researchers, for example, Ngwe and colleagues [30] reported that they obtained IC50 values of the 95% ethanolic and water extracts of G. procumbens to be 1.05 μg/mL and 1.06 μg/mL, respectively. Another study reported that the water extract of G. procumbens yielded an IC50 of 0.092 mg/mL [31]. Further investigation from the reported studies leads to the observation of certain parameters that were not applied in our studies. G. procumbens was extracted thrice with water for 12 h, which was different from our extraction protocol. The incubation temperature during the assay also seemed to play an important role in the enzyme activity [32]. Khatib et al. [29] reported low inhibition of the α-glucosidase enzyme using a Momordica charantia ethanolic extract and suggested different origin, maturity, postharvest, and processing conditions as possible factors for their results. On the other hand, this is the first ever reported study on the inhibition of α-glucosidase of C. gynandra ethanolic extracts.
3.4. NO Inhibition Activity of the G. procumbens and C. gynandra Extracts
The G. procumbens 70% and 100% ethanolic extracts had significantly high inhibitory effects on NO production, with IC50 values of 22.91 μg/mL and 25.20 μg/mL, respectively. The C. gynandra extracts also showed remarkable NO production inhibition results. The 70% and 100% ethanolic extracts of C. gynandra have the lowest IC50 values of 66.01 μg/mL and 60.75 μg/mL, respectively, with significant difference. The inhibitory pattern of both herbs was similar in that the inhibitory activity increased gradually from 0% > 20% > 50% > 70% and 100% ethanolic extracts. The MTT assay further confirmed that none of the G. procumbens and C. gynandra extracts possessed cytotoxicity towards the cells, which strengthened the possibility for the development of anti-inflammatory drugs.
3.5. Correlation between TPC, DPPH Scavenging, and NO Inhibition of G. procumbens and C. gynandra Extracts
To justify whether phenolic compounds from the studied herbs correlate with DPPH scavenging and NO inhibitory activity, a Pearson correlation test was performed at the 95% confidence interval. No correlation data were obtained for the α-glucosidase assay since none of the extracts showed any inhibition towards the enzyme. A strong positive or negative correlation has a scored r value between +0.5 and +1.0 or −0.5 and −1.0. If the score is between +0.1 and +0.3 or −0.1 and −0.3, then the association is considered weak [33].
The correlation between TPC and the DPPH scavenging assay showed that the G. procumbens extracts scored a strong r value of 0.8443, whereas C. gynandra scored lower with r = 0.5888. This indicates that phenolic compounds could be one of the key contributors to the DPPH scavenging activity in both herbs. DPPH is a stable free radical and is frequently used to test the antioxidant capacity of herbal extracts. Plants' phenolic acids have been proven to be the main source of DPPH scavenging activity [28] in which DPPH free radicals receive electrons from phenolic acids and are reduced to the stable DPPH-H complex [34]. Our finding is also in line with the data presented for G. procumbens extracts [35]. The Pearson correlation between TPC and the NO inhibition activity of G. procumbens extract showed a moderately strong r value at 0.6418. This indicates a possible contribution from phenolic compounds in the inhibition of NO production via RAW 264.7 cell induction. By contrast, the C. gynandra extracts showed a weak negative correlation with an r value of 0.2432. A weak negative correlation may be due to the TPC assay providing an estimation of the total phenolic compounds presents in an extract. However, nonphenolic compounds, such as ascorbic acid and tocopherol, are also able to reduce Folin–Ciocalteu's reagent [36]. The presence of nonphenolic metabolites in C. gynandra extracts could contribute to a higher TPC and may not inhibit NO production.
3.6. Tentative Identification and Characterization of Phenolic Compounds from the G. procumbens and C. gynandra Extracts
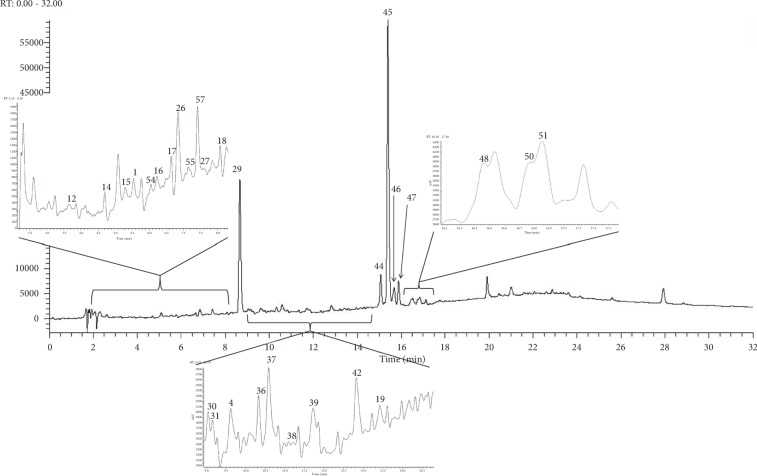
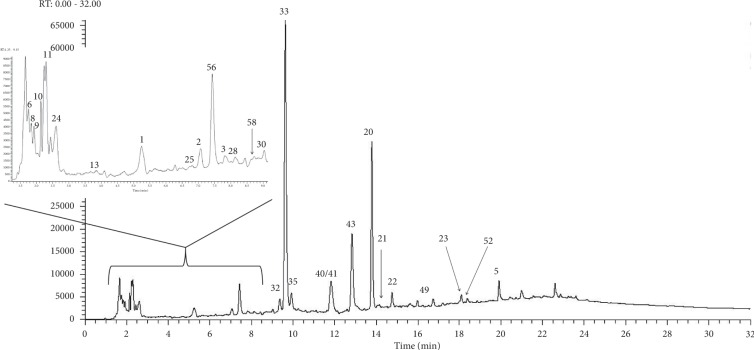
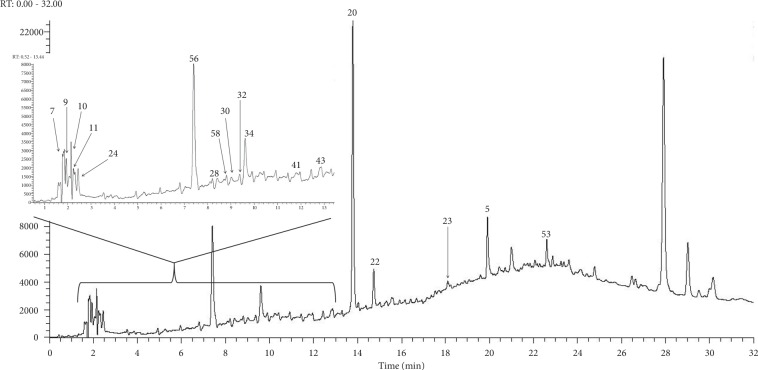
The extracts with the best representation of their total phenolic contents, DPPH scavenging activities, α-glucosidase inhibition, and nitric oxide (NO) inhibition from both herbs were chosen for analysis by UHPLC-ESI-Orbitrap-MS. Ergo, the 100% ethanolic extract of G. procumbens and the 20% and 100% ethanolic extracts of C. gynandra were chosen for this purpose. Table 2 shows the list of tentatively identified phenolic acids grouped according to their class. Among the 58 phenolic acids identified, 27 were found only in the G. procumbens extract, and 11 and 3 were found only in the 20% and 100% ethanolic extracts of C. gynandra, respectively. The remaining 17 phenolic acids can be found in at least two of the extracts analysed. The theoretical monoisotopic mass and mass error of phenolic acids, which were identified as derivative or dimer, were not determined since information of their actual structure were lacking. This analysis also showed that hydroxycinnamic acids were the major phenolic acids identified in all extracts. Other classes of phenolic acids were also identified, including the hydroxybenzoic acids, flavonoid derivatives, organic acids, and sugar derivatives. Figures 1–3 show the chromatograms of the 100% ethanolic extract of G. procumbens and the 20% and 100% ethanolic extracts of C. gynandra, respectively.
Table 2.
Tentative identification of phenolic compounds from the extracts of G. procumbens and C. gynandra.
| No | T R (min) | Measured mass[M-H] − m/z | Theoretical mass[M-H] − m/z | Mass error (ppm) | MS/MSm/z (% intensity) | Tentatively identified metabolites | 100% GP | 20% CG | 100% CG | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| Sugar derivatives | ||||||||||
| 1 | 5.55 | 371.0621 | 371.0693 | −19.4 | 209 (20), 191 (20), 179 (5), 173 (5), 85 (100) | Caffeoylglucaric acid | + | + | − | [37] |
| 2 | 7.07 | 355.0671 | 355.0744 | −20.6 | 209 (4), 191 (3), 163 (4), 119 (5), 85 (100) | Coumaroylglucaric acid | − | + | − | [37] |
| 3 | 7.82 | 355.0673 | 355.0744 | −20.0 | 209 (40), 191 (10), 163 (3), 133 (8), 119 (4), 85 (100) | Isomer of coumaroylglucaric acid | − | + | − | [37] |
| 4 | 9.62 | 355.0673 | 355.0744 | −20.0 | 337 (1), 209 (20), 191 (20), 173 (3), 163 (3), 85 (100) | Isomer of coumaroylglucaric acid | + | − | − | [37] |
| 5 | 19.91 | 465.2257 | n.d | n.d | 261 (6), 243 (100), 221 (70), 177 (3), 149 (2) | Derivative of acetylglucose | − | + | + | [37] |
|
| ||||||||||
| Organic acids | ||||||||||
| 6 | 1.76 | 275.0216 | n.d | n.d | 159 (20), 227 (1), 133 (1), 115 (100), 71 (25) | Derivative of malic acid | − | + | − | [38] |
| 7 | 1.78 | 217.9913 | n.d | n.d | 133 (15), 125 (10), 115 (99), 89 (10), 71 (100) | Derivative of malic acid | − | − | + | [38] |
| 8 | 1.83 | 289.0177 | n.d | n.d | 133 (60), 115 (100), 71 (35) | Derivative of malic acid | − | + | − | [38] |
| 9 | 1.93 | 133.0211 | 133.0215 | −3.0 | 115 (40), 71 (100) | Malic acid | − | + | + | [38] |
| 10 | 2.15 | 405.0248 | n.d | n.d | 191 (40), 173 (10), 129 (5), 111 (100), 87 (20) | Derivative of citric acid | − | + | + | [38] |
| 11 | 2.30 | 191.0189 | 191.0270 | −42.4 | 173 (10), 111 (50), 87 (100), 85 (30), 67 (30), 57 (55) | Citric acid | − | + | + | [38] |
|
| ||||||||||
| Hydroxybenzoic acid derivatives | ||||||||||
| 12 | 3.61 | 315.0724 | 315.0794 | −22.2 | 153 (80), 109 (100) | Protocatechuic acid glucoside | + | − | − | [39] |
| 13 | 3.55 | 351.0599 | n.d | n.d | 198 (10), 180 (4), 169 (20), 125 (60), 111 (100) 79 (60) | Derivative of gallic acid | − | + | − | [40] |
| 14 | 4.53 | 331.0672 | 331.0743 | −21.4 | 313 (6), 169 (100), 150 (40), 125 (98) | Gallic acid glucoside | + | − | − | [40] |
| 15 | 5.30 | 315.0724 | 315.0794 | −22.2 | 152 (80), 108 (100) | Isomer of protocatechuic acid glucoside | + | − | − | [39] |
| 16 | 6.23 | 153.0269 | 153.0266 | 1.96 | 123 (10), 109 (80), 108 (100), 97 (10), 91 (15), 81 (10), 65 (13) | Protocatechuic acid | + | − | − | [39] |
| 17 | 6.64 | 211.0607 | n.d | n.d | 153 (60), 148 (100), 138 (12), 136 (15), 123 (10), 120 (20), 109 (70), 108 (40), 95 (20) | Protocatechuic acid derivative | + | − | − | [39] |
| 18 | 8.27 | 299.0772 | 299.0845 | −24.4 | 137 (60), 93 (100) | Hydroxybenzoic acid glucoside | + | − | − | [41] |
| 19 | 13.42 | 280.0649 | n.d | n.d | 217 (6), 145 (40), 137 (20), 119 (100), 117 (15), 93 (15) | Derivative of hydroxybenzoic acid | + | − | − | [41] |
|
| ||||||||||
| Flavonoid derivatives | ||||||||||
| 20 | 13.80 | 609.1464 | 609.1534 | −11.5 | 343 (4), 301 (100), 179 (5), 151 (4) | Quercetin rutinoside | − | + | + | [38] |
| 21 | 14.28 | 463.0884 | 463.0877 | 1.5 | 301 (50), 300 (100), 271 (5), 255 (3), 179 (4), 175 (2), 151 (4) | Quercetin glucoside | − | + | − | [38] |
| 22 | 14.74 | 593.1516 | 593.1585 | −11.6 | 327 (3), 285 (100), 151 (2) | Kaempferol rutinoside | − | + | + | [38] |
| 23 | 18.26 | 615.2463 | n.d | n.d | 571 (10), 553 (6), 386 (4), 285 (100), 241 (4), 213 (1) | Derivative of luteolin | − | + | + | [38] |
|
| ||||||||||
| Hydroxycinnamic acid derivatives | ||||||||||
| 24 | 2.44 | 248.0539 | n.d | n.d | 180 (20), 163 (60), 153 (10), 119 (100), 107 (15), 93 (20), 72 (50) | Derivative of coumaric acid | − | + | + | [41] |
| 25 | 6.80 | 341.0881 | 341.0951 | −20.5 | 179 (70), 135 (100) | Caffeic acid glucoside | − | + | − | [42] |
| 26 | 6.85 | 353.0879 | 353.0951 | −20.4 | 191 (100), 179 (50), 161 (4), 135 (80), 111 (4), 93 (2), 85 (4) | 3-Caffeoylquinic acid | + | − | − | [38, 41, 42] |
| 27 | 7.56 | 385.0718 | 385.0713 | 1.29 | 223 (40), 205 (60), 147 (10), 129 (30), 111 (60), 101 (20), 85 (100) | Sinapic acid glucoside | + | − | − | [43] |
| 28 | 8.13 | 297.0616 | n.d | n.d | 179 (1), 161 (10), 135 (100), 117 (10), 89 (20) | Derivative of caffeic acid | − | + | + | [42] |
| 29 | 8.66 | 707.1831 | n.d | n.d | 353 (20), 191 (100), 179 (1), 161 (1) | Dimer of caffeoylquinic acid | + | − | − | [44] |
| 30 | 9.04 | 325.0930 | 325.1002 | −22.1 | 163 (20), 119 (100) | Coumaric acid glucoside | + | + | + | [41] |
| 31 | 9.16 | 353.0879 | 353.0951 | −20.4 | 191 (80), 179 (50), 173 (70), 161 (10), 135 (100), 111 (20), 93 (30) | 4-Caffeoylquinic acid | + | − | − | [41] |
| 32 | 9.37 | 369.0369 | 369.0365 | 1.08 | 207 (10), 189 (25), 127 (100), 83 (50) | Dimethoxycinnamoyl glucoside | − | + | + | [45] |
| 33 | 9.63 | 739.1000 | n.d | n.d | 369 (12), 207 (15), 189 (100), 127 (20) | Dimer of dimethoxycinnamoyl glucoside | − | + | − | [45] |
| 34 | 9.63 | 369.0364 | 369.0365 | −0.27 | 207 (2), 189 (10), 127 (100), 99 (20), 83 (80) | Dimethoxycinnamoyl glucoside | − | − | + | [45] |
| 35 | 9.92 | 179.0341 | 179.0423 | −45.8 | 135 (100), 117 (8), 107 (10), 89 (15) | Caffeic acid | − | + | − | [37, 42] |
| 36 | 10.33 | 353.0879 | 353.0951 | −20.4 | 191 (100), 179 (1), 173 (2), 161 (3), 127 (2), 111 (1), 85 (4) | 5-Caffeoylquinic acid | + | − | − | [38, 41, 42] |
| 37 | 10.58 | 337.0931 | 337.1002 | −21.1 | 191 (100), 173 (5), 163 (20), 127 (3), 119 (20), 111 (10), 93 (50) | trans-5-p-Coumaroylquinic acid | + | − | − | [46] |
| 38 | 11.09 | 321.1015 | n.d | n.d | 321 (20), 241 (2), 175 (2), 147 (20), 119 (2), 103 (4), 97 (100) | Derivative of cinnamic acid | + | − | − | [47] |
| 39 | 11.71 | 367.1034 | 367.1107 | −19.9 | 191 (100), 173 (30), 155 (4), 134 (20), 111 (20), 93 (50) | Feruloylquinic acid | + | − | − | [38, 46] |
| 40 | 11.83 | 375.0332 | n.d | n.d | 185 (50), 163 (30), 134 (10), 127 (25), 119 (100), 103 (10), 101 (40), 83 (20) | Derivative of coumaric acid | − | + | − | [41] |
| 41 | 11.83 | 353.0409 | 353.0404 | 1.41 | 207 (1), 189 (10), 127 (100), 119 (5), 99 (20), 83 (90) | Dimethoxycinnamoyl coumaric acid | − | + | + | [45] |
| 42 | 12.82 | 337.0930 | 337.1002 | −21.4 | 191 (100), 173 (2), 163 (2), 127 (2), 119 (2), 93 (4) | cis-5-p-Coumaroylquinic acid | + | − | − | [38, 46] |
| 43 | 12.84 | 383.0620 | 383.0736 | −30.3 | 207 (1), 189 (10), 127 (100), 99 (20), 83 (80) | Dimethoxycinnamoyl glucoronide | − | + | + | [45] |
| 44 | 15.07 | 515.1193 | 515.1267 | −14.4 | 353 (30), 335 (20), 191 (40), 179 (90), 173 (100), 161 (20), 135 (20) | 3,4-Dicaffeoylquinic acid | + | − | − | [48] |
| 45 | 15.38 | 515.1195 | 515.1267 | −14.4 | 353 (20), 191 (100), 179 (75), 173 (4), 161 (4), 135 (10) | 3,5-Dicaffeoylquinic acid | + | − | − | [49] |
| 46 | 15.66 | 515.1196 | 515.1267 | −14.2 | 353 (20), 191 (100), 179 (80), 173 (15), 161 (5), 135 (10) | 1,5-Dicaffeoylquinic acid | + | − | − | [49] |
| 47 | 15.86 | 515.1196 | 515.1267 | −14.2 | 353 (25), 191 (30), 179 (80), 173 (100), 161 (2), 155 (6), 135 (10) | 1,4-Dicaffeoylquinic acid | + | − | − | [48] |
| 48 | 16.43 | 499.1019 | 499.1002 | 3.4 | 337 (3), 191 (8), 173 (6), 163 (100), 119 (6) | Caffeoyl-coumaroylquinic acid | + | − | − | [50] |
| 49 | 16.75 | 387.1086 | n.d | n.d | 193 (100), 179 (3), 161 (8), 133 (4) | Dimer of caffeic acid methyl ester | − | + | − | [42, 51] |
| 50 | 16.77 | 529.1353 | 529.1424 | −13.4 | 367 (8), 193 (100), 179 (4), 173 (8) | Caffeoyl-feruloylquinic acid | + | − | − | [40] |
| 51 | 16.85 | 515.1196 | 515.1267 | −14.2 | 353 (30), 191 (40), 179 (80), 173 (100), 155 (10), 135 (10) | 4,5-Dicaffeoylquinic acid | + | − | − | [48] |
| 52 | 18.39 | 207.0657 | 207.0736 | −38.2 | 179 (2), 161 (30), 135 (90), 133 (100), 117 (2), 106 (3), 89 (2) | Caffeic acid ethyl ester | − | + | − | [42, 51] |
| 53 | 22.61 | 559.3124 | n.d | n.d | 339 (3), 277 (100), 253 (20), 235 (2), 179 (1), 161 (2) | Derivative of caffeic acid | − | − | + | [42] |
|
| ||||||||||
| Other classes of phenolic acid | ||||||||||
| 54 | 6.04 | 315.1087 | 315.1158 | −22.5 | 153 (60), 123 (100) | Hydroxytyrosol glucoside | + | − | − | [39] |
| 55 | 7.03 | 403.0915 | n.d | n.d | 343 (30), 241 (20), 181 (4), 166 (3), 151 (6), 139 (4), 111 (80), 97 (100) | Derivative of syringaldehyde | + | − | − | [39] |
| 56 | 7.40 | 339.0721 | 339.0794 | −21.5 | 225 (2), 203 (2), 177 (100), 133 (10) | Esculetin glucoside | − | + | + | [52] |
| 57 | 7.42 | 471.1146 | 471.1217 | −15.1 | 177 (100), 133 (10) | Esculetin sambubioside | + | − | − | [52] |
| 58 | 8.72 | 343.1035 | 343.0906 | −37.6 | 181 (100), 137 (70), 121 (8), 109 (10) | Homovanillic acid glucoside | − | + | + | [38] |
GP: Gynura procumbens, CG: Cleome gynandra, n.d: not determined. (−) and (+) indicate absence or presence of the metabolite in the extract. Bold metabolites indicate metabolites identified tentatively for the first time in the respective herbs.
Figure 1.
UHPLC chromatogram of 100% ethanolic extract of G. procumbens at 280 nm.
Figure 2.
UHPLC chromatogram of 20% ethanolic extract of C. gynandra at 280 nm.
Figure 3.
UHPLC chromatogram of 100% ethanolic extract of C. gynandra at 280 nm.
3.6.1. Hydroxycinnamic Acid Derivatives
A total of 30 phenolic acids within the class of hydroxycinnamic acid and its derivatives were tentatively identified from all extracts. Metabolites 26, 31, and 36 were found to have a precursor ion at m/z 353 [M-H]−, further fragmentations yielded product ions at m/z 191, 179, 173, and 135, which was similar to the ionization pattern of caffeoylquinic acid [38, 41]. The existence of isomers in caffeoylquinic acid has been well documented based on the difference in intensity of the product ions m/z 179 and 173. Metabolite 31 had an intense signal at m/z 173 due to the loss of [quinic acid-H-H2O]−, which was lacking in metabolites 26 and 36 due to the particular stereochemical arrangement of their structures. 4-Caffeoylquinic acid was confirmed to show such fragmentation patterns. Metabolites 26 and 36 were distinguished from each other based on their retention time reported from past study and assigned as 3-caffeoylquinic acid and 5-2caffeoylquinic acid, respectively [42]. Metabolite 29 with precursor ion m/z 707 [M-H]− was observed as one of the major peaks in the 100% ethanolic extract of G. procumbens; it produced product ions similar to caffeoylquinic acid but with a dimeric adduct of itself as described in [44]. As such, metabolite 29 was assigned as a dimer of caffeoylquinic acid.
Metabolites 44-47 and 51 shared a similar precursor ion at m/z 515 [M-H]− with a base peak at m/z 353 and common product ions at m/z 191, 179, 173, 161, and 135. These fragmentation patterns are similar to dicaffeoylquinic acid. Further investigation of MS3 revealed that metabolites 44, 47, and 51 have m/z 173 [quinic acid-H-H2O]− as the base peak, which indicates the presence of a caffeoyl moiety on the quinic acid at position C-4. The caffeoyl moiety attached at a C position other than C-4 in quinic acid will give a base peak at m/z 191 [quinic acid]− as shown in metabolites 45 and 46. Based on the retention time shift in the reverse phase chromatogram, metabolites 44, 47, and 51 were assigned as 3,4-dicaffeoylquinic acid, 1,4-caffeoylquinic acid, and 4,5-dicaffeoylquinic acid, respectively [48]. Metabolites 45 and 46 were named 3,5-dicaffeoylquinic acid and 1,5-dicaffeoylquinic acid, respectively, since both metabolites eluted just after 3,4-dicaffeoylquinic acid on a reverse-phase column [49]. Comparing the two metabolites, metabolite 45 is structurally more polar than metabolite 46, which makes the former elute earlier as well [49].
Metabolites 24, 30, 37, 40, 42, and 48 have a loss of either coumaric acid (164 amu) or a coumaroyl moiety (146 amu) in their fragmentations, which makes identification easier. Metabolite 30 has a precursor ion m/z 325 [M-H]− with product ion at m/z 163 [M-H-glucose]− and 119 [M-H-glucose-CO2]− which was confirmed as coumaric acid glucoside [41]. Metabolites 37 and 42 with the deprotonated ion m/z 337 [M-H]− produced a base peak ion at m/z 191 [M-H-coumaroyl]−, which indicates the presence of quinic acid. Subsequent MS2 fragmentations produced ions m/z 173 [M-H-coumaroyl-H2O]− and m/z 127, which confirmed the identity of metabolites 37 and 42 as coumaroylquinic acid [38, 46]. 5-p-coumaroylquinic acid has a unique fragmentation with m/z 191 as the base peak compared to other coumaroylquinic acid isomers. Since the report [46] was in accordance with our finding, metabolites 37 and 42 were named trans-5-p-coumaroylquinic acid and cis-5-p-coumaroylqunic acid, respectively. Metabolite 48 adds an additional caffeoyl moiety onto coumaroylquinic acid, which gave the precursor ion m/z 499 [M-H]−. This metabolite was assigned as caffeoyl-coumaroylquinic acid [50]. Metabolites 24 and 40 were assigned as derivatives of coumaric acid since they have unknown adducts on their coumaric acid structure [41].
Metabolite 35 was identified as caffeic acid with precursor ion m/z 179 [M-H]− and product ions at m/z 135 [M-H-44]− and 117 [M-H-44-18]− after the loss of a carbon dioxide and a water molecule [42]. The structures of metabolites 25, 28, 49, 52, and 53 were all associated with caffeic acid. Metabolite 25 loses a glucose moiety from its aglycon and is named caffeic acid glucoside [37, 42]. Metabolite 49 with precursor ion m/z 387 [M-H]− has a base peak at m/z 193, which suggests that it is a dimer. Further MS2 data produced product ion m/z 179 [193-CH3]− after the elimination of a methyl group. As such, metabolite 49 was labelled as a dimer of caffeic acid methyl ester. The presence of caffeic acid ethyl ester was confirmed in metabolite 52 after the precursor ion m/z 207 [M-H]− produced product ion m/z 179 [M-H-29]− due to the elimination of an ethyl moiety. Notably, although both metabolites 49 and 52 were tentatively identified for the first time in C. gynandra, the existence of other varieties of caffeic acid esters in herbs also suggests the possible occurrence of metabolites 49 and 52 [51]. Metabolites 28 and 53 were noted as derivatives of caffeic acid since the conjugation of unknown moieties in caffeic acid produced precursor ions m/z 297 [M-H]− and 599 [M-H]−, respectively.
Metabolites 32-34, 41, and 43 were linked to dimethoxycinnamoyl as they shared the same product ions at m/z 207 [M-H]−, 189 [M-H-H2O]−, 127 [M-H-H2O-2(OCH3)]−, and 83 [M-H-H2O-2(OCH3)-CO2]− [37]. Metabolites 32 and 34 have a glucose moiety in their structure that gave the precursor ion m/z 369 [M-H]−. Metabolite 33 with precursor ion m/z 739 [M-H]− indicates a dimeric molecule of dimethoxycinnamoyl glucoside. Metabolites 41 and 43 with precursor ions m/z 353 [M-H]− and 383 [M-H]−, respectively, suggested the addition of a coumaroyl unit (146 amu) and glucuronide (176 amu) in their dimethoxycinnamoyl structure. MS2 spectra of metabolite 41 produced product ion m/z 119 [163-CO2]−, which further rectified this finding. Other hydroxycinnamic acid derivatives, including sinapic acid glucoside (27), feruloylquinic acid (39), caffeoyl-feruloylquinic acid (50), and derivative of cinnamic acid (38) were identified based on previously reported data [38, 40, 43, 46, 47].
3.6.2. Hydroxybenzoic Acid Derivatives
Protocatechuic acid m/z 153 [M-H]− was associated with metabolites 12, 15, and 17 with metabolite 16 identified as protocatechuic acid and had a base peak at m/z 109 [M-H-CO2]− after elimination of a carbon dioxide molecule. Metabolites 12 and 15 are isomers with precursor ion m/z 315 [M-H]−. Elimination of a sugar molecule (162 amu) from the precursor ion produced a base peak similar to that of protocatechuic acid, m/z 153 [M-H]−. As such, metabolites 12 and 15 were labelled as protocatechuic acid glucoside [39]. Metabolite 17 has a deprotonated m/z 211 [M-H]− with a base peak of 148 [M-H-63]−, indicating a loss of an unknown moiety; probably a methoxy (-OCH3) and an oxygen molecule. The same precursor ion has a product ion at m/z 153 [M-H-58]−, which could be a loss of a methyl formate moiety (-CO(OCH3)), and the MS2 data showed m/z 109 [M-H-58-CO2]−, similar to protocatechuic acid. Due to the uncertainty of the nature of the adduct, metabolite 17 was assigned as a protocatechuic acid derivative.
Metabolite 13 has a deprotonated ion at m/z 351 [M-H]− and ion fragments at m/z 169 after the expulsion of an unknown moiety (182 amu), followed by m/z 125 [M-H-182-CO2]−. Metabolite 14, on the other hand, has a precursor ion m/z 331 [M-H]− with base peak m/z 169 [M-H-glucose]− and MS2 fragmentation ion 125 [M-H-glucose-CO2]−. Based on the mass fragmentation pattern, metabolites 13 and 14 were assigned as derivatives of gallic acid and gallic acid glucoside, respectively [40]. Metabolite 18 has a deprotonated ion m/z 299 [M-H]− and product ion m/z 137 [M-H-162]− and base peak m/z 93 [M-H-162-CO2]−, which confirmed it as hydroxybenzoic acid glucoside [41]. Metabolite 19 has the precursor ion m/z 280 and base peak at m/z 119 [M-H-161]−. Other product ions of metabolite 19 include m/z 137 [M-H-143]− and 93 [137-CO2]−, which represent the possible structure of a hydroxybenzoic acid. As such, metabolite 19 was labelled as a derivative of hydroxybenzoic acid.
3.6.3. Flavonoid Derivatives
Metabolites 20–23 were clustered under the flavonoid derivatives. Metabolite 20 has a deprotonated ion at m/z 609 [M-H]− with a base peak m/z 301 [M-H-308]− after the elimination of a rutinose moiety. The MS2 spectra of m/z 301 produced m/z 179 [M-H-308-122]− and 151 [M-H-308-122–28]−, which are in line with quercetin ion fragmentations. Thus, metabolite 20 was identified as quercetin rutinoside. Quercetin glucoside was identified as metabolite 21 since it has the precursor ion m/z 463 [M-H]− and a base peak similar to the quercetin ion after the elimination of a glucose molecule. Metabolite 22 was found to be associated with kaempferol since fragmentation of precursor ion m/z 593 [M-H]− produced base peak m/z 285 [M-H-308]− and ion m/z 151, which is similar to kaempferol rutinoside [38]. Metabolite 23 has a base peak at m/z 285 [M-H]− after the loss of an unknown mass (330 amu) from the precursor ion m/z 615 [M-H]−. The MS2 spectra of base peak m/z 285 produced ions at m/z 241 [M-H-44]− and 213 [M-H-44-28]−, which are identical to the ions of luteolin [38]. Ergo, metabolite 23 was named as a derivative of luteolin.
3.6.4. Sugar Derivatives
Glucaric acid, m/z 209 [M-H]−, and its ion fragmentations at m/z 191 [M-H-H2O]− and 85 [M-H-2H2O-2CO2]− were associated with metabolites 1-4 [37]. Metabolite 1 has a caffeoyl moiety conjugated with glucaric acid and is thus identified as caffeoylglucaric acid. In return, metabolites 2-4 have a coumaroyl moiety conjugated in their structure and are labelled coumaroylglucaric acid and an isomer of coumaroylglucaric acid, respectively. Metabolite 5 was identified as a derivative of acetylglucoside since its precursor ion m/z 465 [M-H]− has a base peak at m/z 243 after a possible elimination of the acetylglucose moiety (222 amu). Further ion fragmentations produced m/z 221 [M-H-222-22]− and 177 [M-H-222-22-CO2]−, which remains unknown.
3.6.5. Organic Acids
Metabolites 6-11 were identified as malic acid, citric acid, and their derivatives, respectively, based on their fragmentation spectra. Malic acid, m/z 133 [M-H]−, had product ions at m/z 115 [M-H-H2O]− and 71 [M-H-H2O-CO2]−, whereas citric acid, m/z 191 [M-H]− with product ions m/z 173 [M-H-H2O]− and 111 [M-H-2H2O-CO2]−, confirmed their identity [38].
3.6.6. Other Classes of Phenolic Compounds
Other classes of phenolic compounds such as dihydroxybenzene (hydroxytyrosol glucoside 54), hydroxybenzaldehyde (derivative of syringaldehyde 55), hydroxycoumarin (esculetin glucoside 56, esculetin sambubioside 57), and hydroxyphenylacetic acid (homovanillic acid glucoside 58) were tentatively identified based on previously reported data [38, 39, 52].
4. Conclusions
The DPPH scavenging, α-glucosidase inhibition, and NO inhibition activities as well as TPC were tested for G. procumbens and C. gynandra extracted with different ethanolic concentrations. The results showed a preliminary understanding of the potential of both herbs to serve as antioxidant and anti-inflammatory agents. In total, 58 metabolites were identified in both herbs with 24 metabolites were identified for the first time. Tentatively identified metabolites help to reduce the gap in unknown metabolites from both herbs and are important for future reference. Future studies using both herbs as herbal formulations should investigate the synergistic effects of the metabolites on herbal product development.
Acknowledgments
This research was funded by the Malaysian Agricultural Research and Development Institute (MARDI) under RMK-10 fund (P-RBH03-1001).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Iskander M. N., Song Y., Coupar I. M., Jiratchariyakul W. Antiinflammatory screening of the medicinal plant Gynura procumbens. Plant Foods for Human Nutrition. 2002;57(3/4):233–244. doi: 10.1023/a:1021851230890. [DOI] [PubMed] [Google Scholar]
- 2.Wiart C. Medicinal Plants of Asia and the Pacific. 1st. New York, NY, USA: Taylor & Francis Group; 2006. [Google Scholar]
- 3.Abrika O. S. S., Yam M. F., Asmawi M. Z., Sadikun A., Dieng H., Hussain E. A. Effects of extracts and fractions of Gynura procumbens on rat atrial contraction. Journal of Acupuncture and Meridian Studies. 2013;6(4):199–207. doi: 10.1016/j.jams.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Shwter A. N., Abdullah N. A., Alshawsh M. A., et al. Chemoprevention of colonic aberrant crypt foci by Gynura procumbens in rats. Journal of Ethnopharmacology. 2014;151(3):1194–1201. doi: 10.1016/j.jep.2013.12.044. [DOI] [PubMed] [Google Scholar]
- 5.Zahra A. A., Kadir F. A., Mahmood A. A., et al. Acute toxicity study and wound healing potential of Gynura procumbens leaf extracts in rats. Journal of Medicinal Plants Research. 2011;5:2551–2558. [Google Scholar]
- 6.Kim J., Lee C.-W., Kim E. K., et al. Inhibition effect of Gynura procumbens extract on UV-B-induced matrix-metalloproteinase expression in human dermal fibroblasts. Journal of Ethnopharmacology. 2011;137(1):427–433. doi: 10.1016/j.jep.2011.04.072. [DOI] [PubMed] [Google Scholar]
- 7.Hassan Z., Yam M. F., Ahmad M., Yusof A. P. M. Antidiabetic properties and mechanism of action of Gynura procumbens water extract in streptozotocin-induced diabetic rats. Molecules. 2010;15(12):9008–9023. doi: 10.3390/molecules15129008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mishra S. S., Moharana S. K., Dash M. R. Review on Cleome gynandra. International Journal of Research in Pharmaceutical Sciences. 2011;1:681–689. [Google Scholar]
- 9.Anbazhagi T., Kadavul K., Suguna G., Petrus A. J. A. Studies on the pharmacognostical and in vitro antioxidant potential of Cleome gynandra Linn. leaves. Natural Product Radiance. 2009;8:151–157. [Google Scholar]
- 10.Narendhirakannan R. T., Subramanian S., Kandaswamy M. Anti-inflammatory and lysosomal stability actions of Cleome gynandra L. studied in adjuvant induced arthritic rats. Food and Chemical Toxicology. 2007;45(6):1001–1012. doi: 10.1016/j.fct.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Nirosa T., Sravanthi K., Brahma R. R. Evaluation of antidiabetic activity of Cleome gynandra leaves. Indo American Journal of Pharmaceutical Sciences. 2016;3(5):482–486. [Google Scholar]
- 12.Bala A., Kar B., Haldar P. K., Mazumder U. K., Bera S. Evaluation of anticancer activity of Cleome gynandra on Ehrlich’s Ascites Carcinoma treated mice. Journal of Ethnopharmacology. 2010;129(1):131–134. doi: 10.1016/j.jep.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Moyo M., Amoo S. O., Aremu A. O., et al. Determination of mineral constituents, phytochemicals and antioxidant qualities of Cleome gynandra, compared to Brassica oleracea and Beta vulgaris. Frontiers in Chemistry. 2018;5:1–9. doi: 10.3389/fchem.2017.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ranjitha J., Madonna S., Michael D., VIjayalakshmi S. Isolation of novel phytoconstituents from the stem part of Cleome gynandra Linn and their antimicrobial activity. International Journal of Phytomedicine. 2014;6:341–345. [Google Scholar]
- 15.Cheenpracha S., Park E.-J., Rostama B., Pezzuto J., Chang L. C. Inhibition of nitric oxide (NO) production in lipopolysaccharide (LPS)-Activated murine macrophage RAW 264.7 cells by the norsesterterpene peroxide, epimuqubilin A. Marine Drugs. 2010;8(3):429–437. doi: 10.3390/md8030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adela R., Nethi S., Bagul P. K., et al. Hyperglycaemia enhances nitric oxide production in diabetes: a study from South Indian patients. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125270.e0125270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J., Zhao S., Yin P., et al. α-Glucosidase inhibitory activity of polyphenols from the burs of castanea mollissima blume. Molecules. 2014;19(6):8373–8386. doi: 10.3390/molecules19068373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawal U., Leong S. W., shaari K., Ismail I. S., Khatib A., Abas F. α-glucosidase inhibitory and antioxidant activities of different Ipomoea aquatica cultivars and LC-MS/MS profiling of the active cultivar. Journal of Food Biochemistry. 2016;41(2):1–8. doi: 10.1111/jfbc.12303. [DOI] [Google Scholar]
- 19.Chaudhury A., Duvoor C., Dendi V. S. R., et al. Clinical review of antidiabetic drugs: implications for type 2 diabetes mellitus management. Frontiers in Endocrinology. 2017;8:1–12. doi: 10.3389/fendo.2017.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locatelli M., Governatori L., Carlucci G., Genovese S., Mollica A., Epifano F. Recent application of analytical methods to phase I and phase II drugs development: a review. Biomedical Chromatography. 2012;26(3):283–300. doi: 10.1002/bmc.1674. [DOI] [PubMed] [Google Scholar]
- 21.Do Q. D., Angkawijaya A. E., Tran-Nguyen P. L., et al. Effect of extraction solvent on total phenol content, total flavonoid content, and antioxidant activity of Limnophila aromatica. Journal of Food and Drug Analysis. 2014;22(3):296–302. doi: 10.1016/j.jfda.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mediani A., Abas F., Khatib A., et al. Phytochemical and biological features of Phyllanthus niruri and Phyllanthus urinaria harvested at different growth stages revealed by 1 H NMR-based metabolomics. Industrial Crops and Products. 2015;77:602–613. doi: 10.1016/j.indcrop.2015.09.036. [DOI] [Google Scholar]
- 23.Lee S. Y., Mediani A., Nur Ashikin A. H., Azliana A. B. S., Abas F. Antioxidant and α-glucosidase inhibitory activities of the leaf and stem of selected traditional medicinal plants. International Food Research Journal. 2014;21:165–172. [Google Scholar]
- 24.Abdul Hamid N. A., Abas F., Ismail I. S., Shaari K., Lajis N. H. Influence of different drying treatments and extraction solvents on the metabolite profile and nitric oxide inhibitory activity of Ajwa Dates. Journal of Food Science. 2015;80(11):2603–2610. doi: 10.1111/1750-3841.13084. [DOI] [PubMed] [Google Scholar]
- 25.Wishart D. S., Feunang Y. D., Marcu A., et al. HMDB 4.0:the human metabolome database for 2018. Nucleic Acids Research. 2018;46(D1):608–617. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Center for Biotechnology Information. U.S. National Library of Medicine. Rockville pike, MD, USA: National Center for Biotechnology Information; 2018. [Google Scholar]
- 27.Devi P. B., Vijayabharathi R., Sathyabama S., Malleshi N. G., Priyadarisini V. B. Health benefits of finger millet (Eleusine coracana L.) polyphenols and dietary fiber: a review. Journal of Food Science and Technology. 2014;51(6):1021–1040. doi: 10.1007/s13197-011-0584-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Widyawati P. S., Budianta T. D. W., Kusuma F. A., Wijaya E. L. Difference of solvent polarity to phytochemical content and antioxidant activity of Pluchea indica Less leaves extracts. International Journal of Pharmacognosy and Phytochemical Sciences. 2014;15:850–855. [Google Scholar]
- 29.Khatib A., Perumal V., Ahmed Q. U., Uzir B. F., Murugesu S. Low inhibition of alpha-glucosidase and xanthine oxidase activities of ethanol extract of Momordica charantia fruit. Journal of Pharmaceutical Negative Results. 2018;8:20–24. [Google Scholar]
- 30.Ngwe H., Win K. C., Kyaw M. M., Zin T. T., Thu K., Maw S. S. Investigation of the bioactive principles and α-glucosidase inhibitory effect of some Myanmar traditional medicinal plants. Proceeding of the 4th AUN/SEED-Net Regional Conference on Biotechnology; January 2012; Thailand. Faculty of Engineering, Chulalongkorn University and Burapha University; [Google Scholar]
- 31.Choi S.-I., Park M. H., Han J.-S. Gynura procumbens extract alleviates postprandial hyperglycemia in diabetic mice. Preventive Nutrition and Food Science. 2016;21(3):181–186. doi: 10.3746/pnf.2016.21.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elya B., Handayani R., Sauriasari R., et al. Antidiabetic activity and phytochemical screening of extracts from Indonesian plants by inhibition of alpha amylase, alpha glucosidase and dipeptidyl peptidase IV. Pakistan Journal of Biological Sciences. 2015;18(6):279–284. doi: 10.3923/pjbs.2015.279.284. [DOI] [Google Scholar]
- 33.Wilson L. T. Statistical correlation 2009. 2019. https://explorable.com/statistical-correlation.
- 34.Lewis M. J. Natural product screening: anti-oxidant screen for extracts. 2012. https://dokumen.tips/documents/153b-natural-product-screening-anti-oxidant-screen-dpph-of-extract-crude-extract1.html.
- 35.Rosidah, Yam M., Sadikun A., Asmawi M. Antioxidant potential of Gynura procumbens. Pharmaceutical Biology. 2008;46(9):616–625. doi: 10.1080/13880200802179642. [DOI] [Google Scholar]
- 36.Laçine A., Erdi K., Yasin A., Zeyneb A., Mustafa K. Free radical scavenging activity, total phenolic content, total antioxidant status, and total oxidant status of endemic Thermopsis turcica. Saudi Journal of Biological Sciences. 2013;3:235–239. doi: 10.1016/j.sjbs.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S., Lin Z., Jiang H., Tong L., Wang H., Chen S. Rapid identification and assignation of the active ingredients in Fufang Banbianlian injection using HPLC-DAD-ESI-IT-TOF-MS. Journal of Chromatographic Science. 2016;54(7):1225–1237. doi: 10.1093/chromsci/bmw055. [DOI] [PubMed] [Google Scholar]
- 38.Karar M. G. E., Kuhnert N. UPLC-ESI-Q-TOF-MS/MS characterization of phenolics from Crataegus monogyna and Crataegus laevigata (Howthorn) leaves, fruits and their herba derived drops (Crataegutt Trofen) Journal of Chemical Biology & Therapeutics. 2016;1(2):1–23. doi: 10.4172/2572-0406.1000102. [DOI] [Google Scholar]
- 39.Sanz M., Simón B. F., Cadahía E., et al. LC-DAD/ESI-MS/MS study of phenolic compounds in ash (Fraxinus excelsior L. and F. americana L.) heartwood. Effect of toasting intensity at cooperage. Journal of Mass Spectrometry. 2012;47(7):905–918. doi: 10.1002/jms.3040. [DOI] [PubMed] [Google Scholar]
- 40.Spinola V., Pinto J., Catilho P. C. Identification and quantification of phenolic compounds of selected fruits from Madeira island by HPLC-DAD-ESI-MSn and screening for their antioxidant activity. Food Chemistry. 2015;173:14–30. doi: 10.1016/j.foodchem.2014.09.163. [DOI] [PubMed] [Google Scholar]
- 41.Fang N., Yu S., Prior R. L. LC/MS/MS characterization of phenolic constituents in dried plums. Journal of Agricultural and Food Chemistry. 2002;50(12):3579–3585. doi: 10.1021/jf0201327. [DOI] [PubMed] [Google Scholar]
- 42.Ncube E. N., Mhlongo M., Piater L. A., Steenkamp P. A., Dubery I. A., Madala N. E. Analyses of chlorogenic acids and related cinnamic acid derivatives from Nicotiana tabacum tissues with the aid of UPLC-QTOF-MS/MS based on the in-source collision-induced dissociation method. Chemistry Central Journal. 2014;8(1):1–10. doi: 10.1186/s13065-014-0066-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J., Chen M., Ju W., et al. Liquid chromatography/tandem mass spectrometry assay for the simultaneous determination of chlorogenic acid and cinnamic acid in plasma and its application to a pharmacokinetic study. Journal of Pharmaceutical and Biomedical Analysis. 2010;51(3):685–690. doi: 10.1016/j.jpba.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 44.Zwyrzykowska A., Kupczynski R., Jarosz B., Szumny A., Kucharska A. Z. Qualitative and quantitative analysis of polyphenolic compounds in Ilex Sp. Open Chemistry. 2015;13(1):1303–1312. doi: 10.1515/chem-2015-0142. [DOI] [Google Scholar]
- 45.Barros L., Dueñas M., Dias M. I., Sousa M. J., Santos-Buelga C., Ferreira I. C. F. R. Phenolic profiles of in vivo and in vitro grown Coriandrum sativum L. Food Chemistry. 2012;132(2):841–848. doi: 10.1016/j.foodchem.2011.11.048. [DOI] [Google Scholar]
- 46.Chen F., Long X., Liu Z., Shao H., Liu L. Analysis of phenolic acids of jerusalem artichoke (helianthus tuberosusL.) responding to salt-stress by liquid chromatography/tandem mass spectrometry. The Scientific World Journal. 2014;2014:8. doi: 10.1155/2014/568043.568043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simirgiotis M., Benites J., Areche C., Sepúlveda B. Antioxidant capacities and analysis of phenolic compounds in three endemic Nolana species by HPLC-PDA-ESI-MS. Molecules. 2015;20(6):11490–11507. doi: 10.3390/molecules200611490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clifford M. N., Knight S., Kuhnert N. Discriminating between the six isomers of dicaffeoylquinic acid by LC-MSn. Journal of Agricultural and Food Chemistry. 2005;53(10):3821–3832. doi: 10.1021/jf050046h. [DOI] [PubMed] [Google Scholar]
- 49.Tolonen A., Joutsamo T., Mattlla S., Kämäräinen T., Jalonen J. Identification of isomeric dicaffeoylquinic acids fromEleutherococcus senticosususing HPLC-ESI/TOF/MS and1H-NMR methods. Phytochemical Analysis. 2002;13(6):316–328. doi: 10.1002/pca.663. [DOI] [PubMed] [Google Scholar]
- 50.Martucci M. E. O., De Vos R. C. H., Corollo C. A., Neto L. G. Metabolomics as a potential chemotaxonomical tool: application in the genus Vernonia Schreb. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093149.e93149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balachandran C., Emi N., Arun Y., et al. In vitro anticancer activity of methyl caffeate isolated from Solanum torvum Swartz. fruit. Chemico-Biological Interactions. 2015;242:81–90. doi: 10.1016/j.cbi.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 52.Yun E. S., Park S. K., Kim B. S., et al. Determination of the esculetin contents of medicinal plants by liquid chromatography-tandem mass spectrometry. Biomedical Chromatography. 2012;26(10):1247–1251. doi: 10.1002/bmc.2686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.