Figure 4. Su(var)2–10 genetically and biochemically interacts with Piwi, Arx and Panx.
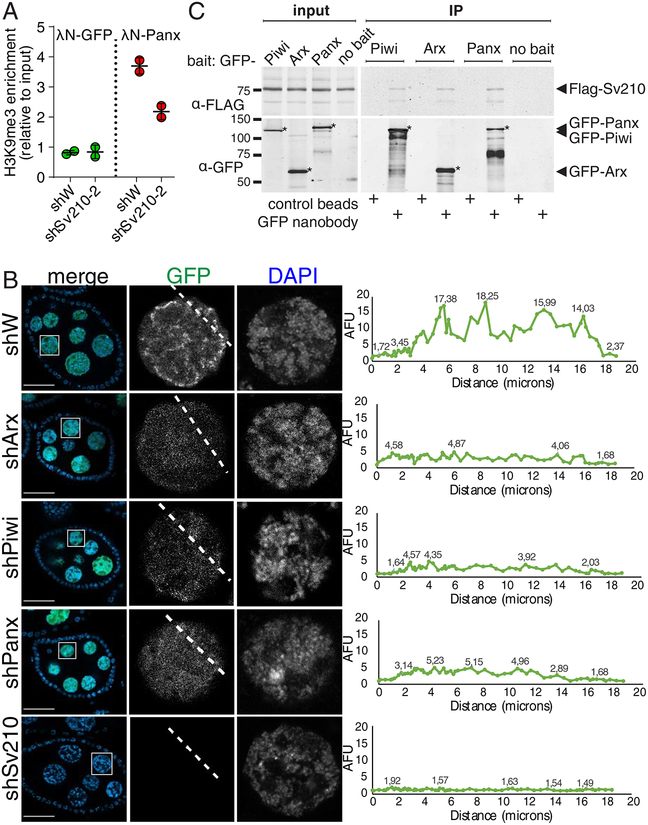
(A) Su(var)2–10 GLKD abolishes H3K9me3 deposition induced by Panx tethering to luciferase reporter (Yu et al., 2015) locus. Plot shows the H3K9me3 enrichment (ChIP-qPCR) at the reporter upon tethering of Panx or GFP control in control (shW) or Su(var)2–10 GLKD ovaries. Dots correspond to two independent biological replicates; bars indicate the mean and SD.
(B) Nuclear localization of Su(var)2–10 depends on Arx, Panx and Piwi. Images show nurse cell nuclei (DAPI; red) of ovaries from flies expressing MT-Gal4-driven GFP-Su(var)2–10 (green) and short hairpins against Asterix (shArx), Piwi (shPiwi), Panoramix (shPanx), Su(var)2–10 (shSv210–2) or white (shW, control). Scale bar 30 μm.
(C) Su(var)2–10 interacts with Piwi, Arx and Panx. Protein lysates from ovaries expressing MT-Gal4 driven FLAG-Su(var)2–10 and GFP-fusion Piwi (endogenous promoter), Arx or Panx, were used for co-immunoprecipitation using GFP nanotrap or control beads. Ovaries expressing FLAG-Su(var)2–10 but no GFP partner (no bait) were used as an additional negative control.