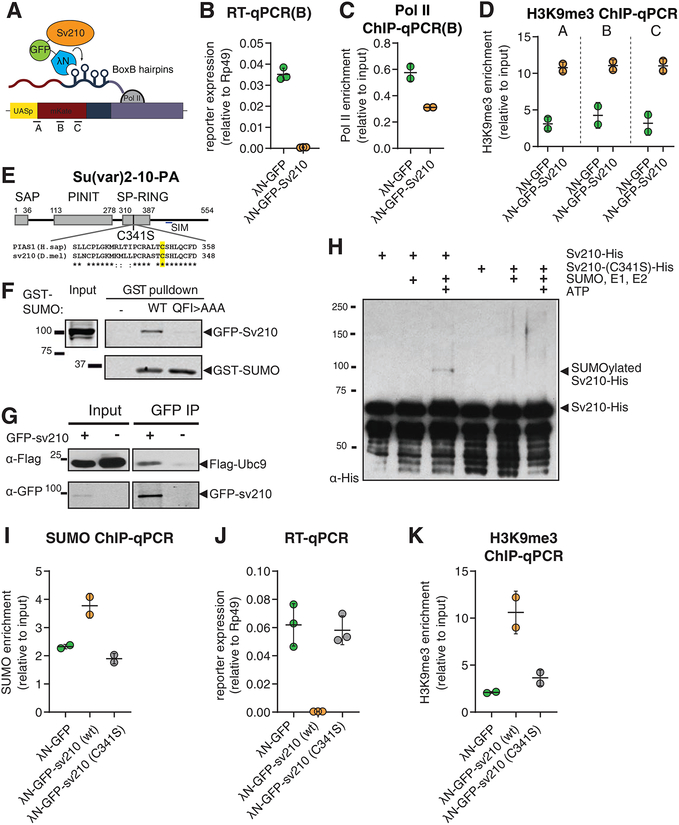
Figure 5. Su(var)2–10 recruitment induces transcriptional repression that depends on the SUMO pathway.
(A) Schematic diagram of the reporter used to study the effect of Su(var)2–10 recruitment to RNA. GFP-Su(var)2–10 fused to the RNA-biding λN domain, and the mKate reporter encoding 4 BoxB hairpins in the 3’UTR, are co-expressed in germ cells of the ovary (using the MT-Gal4 driver), resulting in Su(var)2–10 recruitment to the reporter’s nascent transcript. A-C denote different amplicons used for qPCR analysis.
(B-D) Tethering of Su(var)2–10 leads to reduced reporter mRNA level and Pol II occupancy, and increased H3K9me3 signal. Plots show reporter expression (B), Pol II occupancy (C), and H3K9me3 enrichment (D) upon tethering of λN-GFP-Su(var)2–10 or λN-GFP control. Dots correspond to independent biological replicates; bars indicate the mean and SD.
(E) Diagram of Drosophila Su(var)2–10 protein structure (PA isoform). Grey boxes mark conserved domains. Alignment of the SP-RING domain between human PIAS1 and Su(var)2–10 is shown, highlighting the catalytic cysteine residue identified in human PIAS1 (Kahyo et al., 2001).
(F) Su(var)2–10 interacts with SUMO. S2 cell lysates expressing GFP-Su(var)2–10 were incubated with bacterially expressed GST-SUMO (wild type), interaction-deficient mutant GST-SUMO (QFI>AAA), or no GST bait control. GST-SUMO was affinity purified using glutathione sepharose beads.
(G) Su(var)2–10 interacts with Ubc9. S2 cell lysates expressing FLAG-Ubc9 and GFP-Su(var)2–10 were immunoprecipitated using GFP nanotrap beads.
(H) Su(var)2–10 is SUMOylated in vitro. Bacterially purified His-Su(var)2–10 or His-Su(var)2–10 C341 mutant were incubated with SUMO, SUMO E1 and E2 ligases in the presence or absence of ATP.
(I) Tethering of Su(var)2–10 promotes SUMO accumulation at reporter locus. Plots show SUMO enrichment at the reporter locus upon tethering of λN-GFP control, wild type λN-GFP-Su(var)2–10 or λN-GFP-Su(var)2–10 C341 mutant estimated by ChIP-qPCR. Dots correspond to two independent biological replicates; bars indicate the mean and SD.
(J, K) Su(var)2–10 mutant (C341S) is unable to repress reporter transcription or induce H3K9me3 deposition. Plots show reporter expression (J) and H3K9me3 enrichment at the reporter locus (K) upon tethering of λN-GFP control, λN-GFP-Su(var)2–10 (wild type) or λN-GFP-Su(var)2–10 C341S mutant. Dots correspond to independent biological replicates; bars indicate the mean and SD.
See also Figure S5.