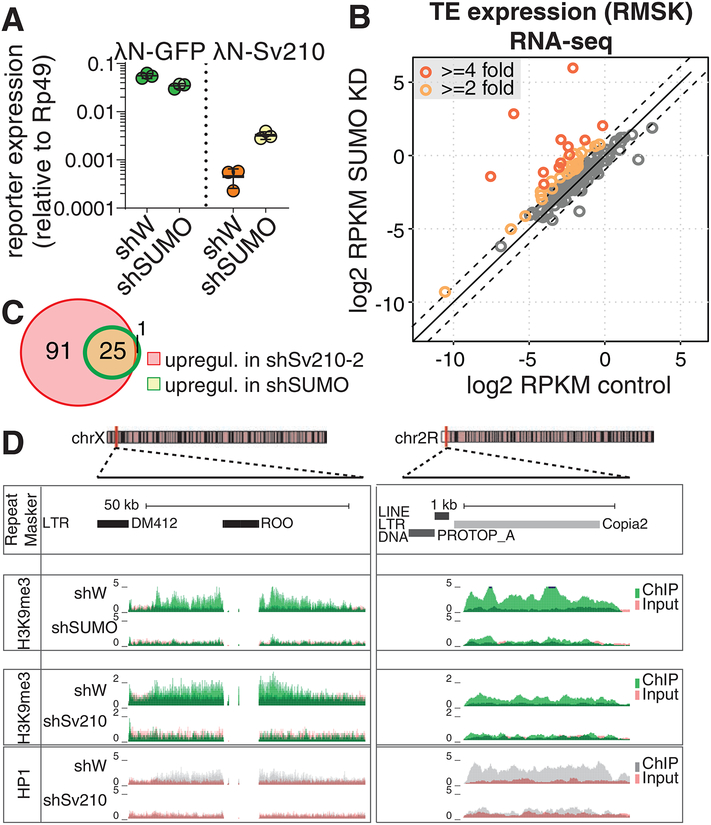
Figure 6. SUMO is required for Su(var)2–10 mediated reporter silencing and global regulation of TEs.
(A) SUMO is required for Su(var)2–10 mediated reporter silencing. Plots show reporter expression upon tethering of λN-GFP or λN-GFP-Su(var)2–10 in SUMO GLKD or control (shW) ovaries measured by RT-qPCR (region B). Dots correspond to three independent biological replicates; bars indicate the mean and SD.
(B) SUMO depletion leads to global transposon de-repression. Scatter plots show log2-tansformed RPKM values for TEs (RepeatMasker annotations) in RNA-seq data from SUMO GLKD versus shW control ovaries. Dashed lines indicate 2-fold change. Data is average from two biological replicates.
(C) Venn diagram shows the numbers and overlap of significantly upregulated TE families upon Su(var)2–10 and SUMO GLKD as determined by DESeq2 (FDR<0.05, log2 fold change ≥2).
(D) SUMO depletion leads to loss of H3K9me3 signal at transposon loci. UCSC browser snapshots showing RPM-normalized H3K9me3 and HP1 ChIP-seq signal (uniquely mapping reads) at TE-rich genomic regions in control (shW), Su(var)2–10 and SUMO GLKD ovaries. ChIP and input signal tracks are overlaid.