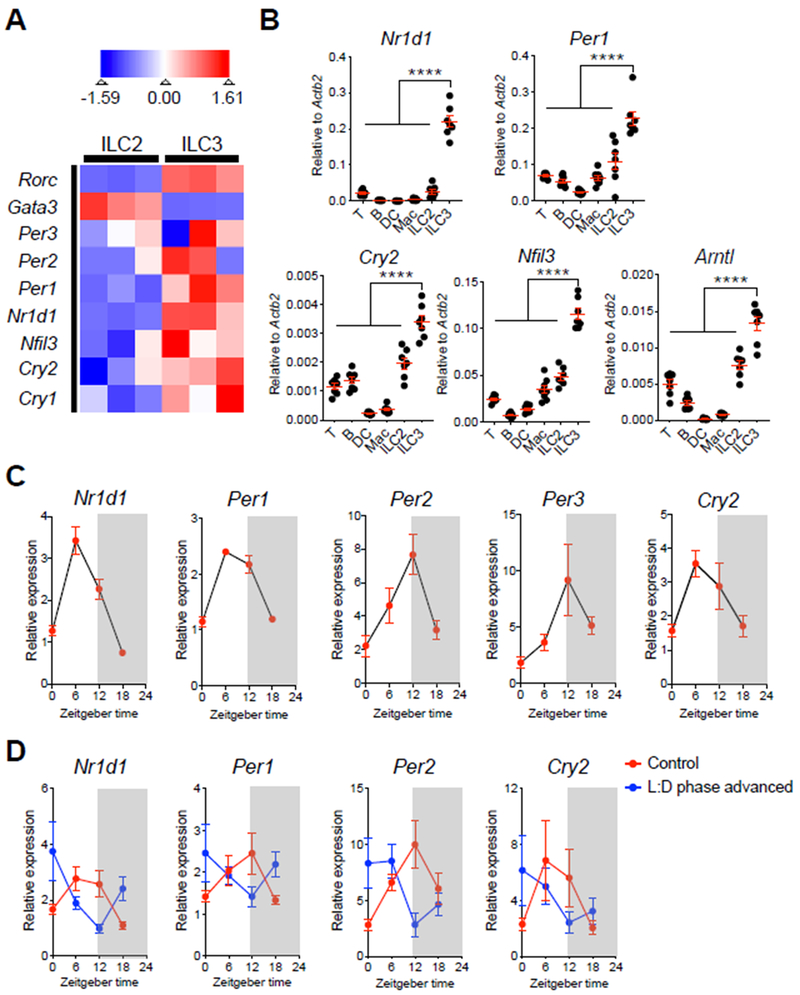
Figure 1. Intestinal ILC3 are enriched for, and exhibit diurnal oscillations in, circadian-related gene expression.
A. Heatmap showing expression Z-scores of the indicated genes in ILC2 (CD45+Lin−CD90.2+CD127+CD27−KLRG1+) and ILC3 (CD45+CD3ε−CD127+RORγtGFP+) from SI-LPs of naïve Rorc(γt)-GfpTG mice, as measured by RNA-sequencing (Lineage markers: CD3ε, CD5, CD8α, NK1.1, CD11c, CD11b, B220). B. qPCR analyses of indicated circadian genes from sort-purified T cells, B cells, dendritic cells (DC), macrophages (Mac), ILC2 and ILC3 from SI-LP of C57BL/6 mice sacrificed at ZT6. The expression of each target gene was normalized to Actb2 (n=7, pooled from 2 independent assays). C. qPCR analyses of diurnal expression patterns in indicated circadian genes on sort-purified ILC3 from SI-LP of C57BL/6 mice sacrificed at ZT0, ZT6, ZT12 and ZT18 within a 24-hour cycle. The expression of each target gene at each time point was first normalized to Actb2, then further normalized to the sample with lowest expression at ZT0 (n=4, representative of 2 independent assays). D. qPCR analyses of diurnal expression patterns in indicated circadian genes on sort-purified ILC3 from SI-LP of C57BL/6 mice housed in standard light:dark cycle (control) or mice housed in 9-hour advanced light:dark cycle (L:D phase advanced). Both groups of mice were sacrificed at ZT0, ZT6, ZT12 and ZT18 within a 24-hour cycle. The expression of each target gene at each time point was first normalized to Actb2, then further normalized to the sample with lowest expression at ZT0 in the control group (n=6/group, pooled from 2 independent assays). Results are shown as the mean ± s.e.m. Statistics are calculated by one-way ANOVA.