Correction to: Human Genome Variation
10.1038/s41439-019-0065-7, published online 2 August 2019
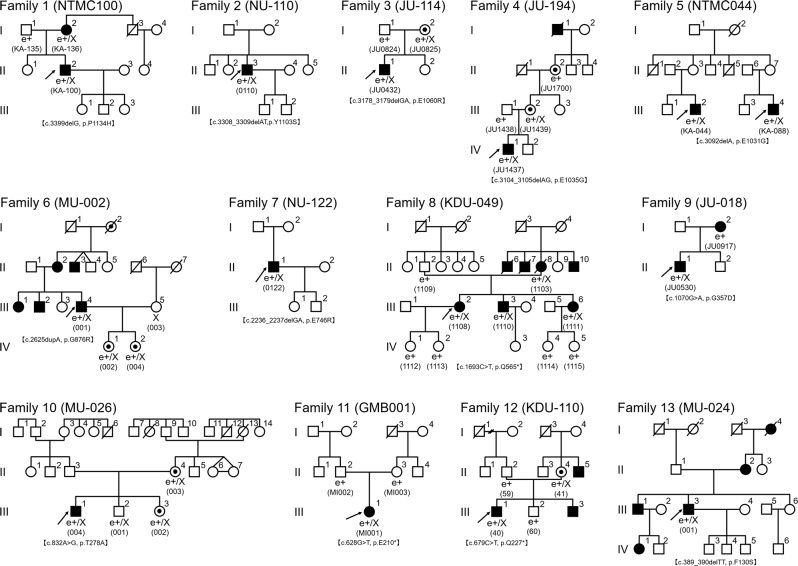
After online publication of this article, the authors noticed an error in the Author name, Results and Discussion section, as well as in Fig. 1 and Table 2.
Fig. 1
Table 2.
Summary of detected variants of 19 affected and 22 unaffected individuals from 13 families with RPGR-associated retinal disorder.
| Fmily No. | Patient No. | Gender | Affected/unaffected | Exon | Nucleotide and amino acid changes | State |
|---|---|---|---|---|---|---|
| 1 | 1-II:2 | Male | Affected | 15 | c.3399delG, p.Pro1134HisfsTer18 | Hemizygous |
| 1-I:1 | Male | Unaffected | ND | |||
| 1-I:2 | Female | Affected | 15 | c.3399delG, p.Pro1134HisfsTer18 | Heterozygous | |
| 2 | 2-II:3 | Male | Affected | 15 | c.3308_3309delAT, p.Tyr1103SerfsTer7 | Hemizygous |
| 3 | 3-II:1 | Male | Affected | 15 | c.3178_3179delGA, p.Glu1060ArgfsTer18 | Hemizygous |
| 3-I:1 | Male | Unaffected | ND | |||
| 3-I:2 | Female | Unaffected | 15 | c.3178_3179delGA, p.Glu1060ArgfsTer18 | Heterozygous | |
| 4 | 4-IV:1 | Male | Affected | 15 | c.3104_3105delAG, p.Glu1035GlyfsTer43 | Hemizygous |
| 4-III:1 | Male | Unaffected | ND | |||
| 4-III:2 | Female | Unaffected | 15 | c.3104_3105delAG, p.Glu1035GlyfsTer43 | Heterozygous | |
| 4-II:2 | Female | Unaffected | ND | |||
| 5 | 5-III:2 | Male | Affected | 15 | c.3092delA, p.Glu1031GlyfsTer58 | Hemizygous |
| 5-III:4 | Male | Affected | 15 | c.3092delA, p.Glu1031GlyfsTer58 | Hemizygous | |
| 6 | 6-III:4 | Male | Affected | 15 | c.2625dupA, p.Gly876ArgfsTer203 | Hemizygous |
| 6-IV:1 | Female | Unaffected | 15 | c.2625dupA, p.Gly876ArgfsTer203 | Heterozygous | |
| 6-III:5 | Female | Unaffected | ND | |||
| 6-IV:2 | Female | Unaffected | 15 | c.2625dupA, p.Gly876ArgfsTer203 | Heterozygous | |
| 7 | 7-II:1 | Male | Affected | 15 | c.2236_2237delGA, p.Glu746ArgfsTer23 | Hemizygous |
| 8 | 8-III:2 | Female | Affected | 14 | c.1693C>T, p.Gln565Ter | Heterozygous |
| 8-II:8 | Female | Affected | 14 | c.1693C>T, p.Gln565Ter | Heterozygous | |
| 8-II:2 | Male | Unaffected | ND | |||
| 8-III:3 | Male | Affected | 14 | c.1693C>T, p.Gln565Ter | Hemizygous | |
| 8-III:6 | Female | Affected | 14 | c.1693C>T, p.Gln565Ter | Heterozygous | |
| 8-IV:1 | Female | Unaffected | ND | |||
| 8-IV:2 | Female | Unaffected | ND | |||
| 8-IV:4 | Female | Unaffected | ND | |||
| 8-IV:5 | Female | Unaffected | ND | |||
| 9 | 9-II:1 | Male | Affected | 10 | c.1070G>A, p.Gly357Asp | Hemizygous |
| 9-I:2 | Female | Affected | ND | |||
| 10 | 10-III:1 | Male | Affected | 8 | c.832A>G, p.Thr278Ala | Hemizygous |
| 10-III:2 | Male | Unaffected | 8 | c.832A>G, p.Thr278Ala | Hemizygous | |
| 10-III:3 | Female | Unaffected | 8 | c.832A>G, p.Thr278Ala | Heterozygous | |
| 10-II:4 | Female | Unaffected | 8 | c.832A>G, p.Thr278Ala | Heterozygous | |
| 11 | 11-III:1 | Female | Affected | 7 | c.628G>T, p.Glu210Ter | Heterozygous |
| 11-II:2 | Male | Unaffected | ND | |||
| 11-II:3 | Female | Unaffected | ND | |||
| 12 | 12-III:1 | Male | Affected | 7 | c.679C>T, p.Gln227Ter | Hemizygous |
| 12-II:4 | Female | Unaffected | 7 | c.679C>T, p.Gln227Ter | Heterozygous | |
| 12-II:2 | Male | Unaffected | ND | |||
| 12-III:2 | Male | Unaffected | ND | |||
| 13 | 13-III:3 | Male | Affected | 5 | c.389_390delTT, p.Phe130SerfsTer4 | Hemizygous |
RPGR transcript ID: NM_001034853.1. Whole-exome sequencing with targeted analysis for retinal disease-causing genes on RetNET (https://sph.uth.edu/retnet/) was performed in 19 affected and 22 unaffected subjects from 13 families. Sequence Variant Nomenclature was obrained according to the guidelines of the Human Genome Variation Society by using Mutalyzer (https://mutalyzer.nl/).
Novel variants are shown in Italic.
ND not detected.
The correct Author name, Results section, Discussion section, Fig. 1, and Table 2 information should have read as follows:
“First, the correct author name is as follows:
Yu Fujinami-Yokokawa.
Second, the number of affected patients (14) refers to the number of probands as mentioned in the title and Results section. The number of carrier patients (7) refers to the carriers regardless of the symptoms as presented in the Results section.
Third, the correct notation symbol for “Family 9-I:2” in Fig. 1 is a solid circle.
All the carriers except Family 9-I:2 were “unaffected” as presented in Table 2. Therefore, the number of affected and unaffected patients were 19 and 22, respectively. The 19 patients affected in Table 2 included 14 probands, 4 carriers with symptoms, and III-3 of Family 8.
Last, the correct notation of the first sentence in the Discussion section is as follows:
The clinical and genetic characteristics of RPGR-RD were illustrated in a nationwide cohort of 15 affected individuals (14 probands and III-3 of Family 8), 14 unaffected individuals, and 12 carriers (4 carriers with symptoms and 8 carriers without symptoms) from 13 Japanese families with RPGR-RD, detecting 13 variants including 8 novel variants.”
The authors apologize for the inconvenience caused.