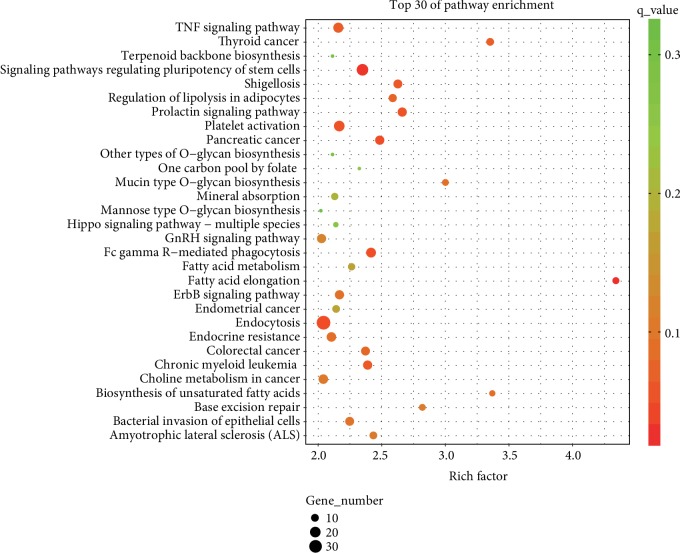
Figure 6.
KEGG pathway analyses of miR-22-3p and miR-320a. The results show the associated target gene pathway of the two miRNAs. The enrichment P values were calculated by Fisher's exact test. Term/pathway in the vertical axis was drawn according to the enrich factor value of the pathway name in descending order. The horizontal axis represents the enrich factor, as follows: enrich factor = (dysregulated gene number in a pathway/total dysregulated gene number)/(gene number in a pathway in the database/total gene number in the database). Top 30 pathway terms were selected according to the enrich factor value. Selection standards included the gene number in a pathway 4 and q < 0.05. Different colors from green to red represent P value. Different sizes of the round shape represent gene count number in a pathway.