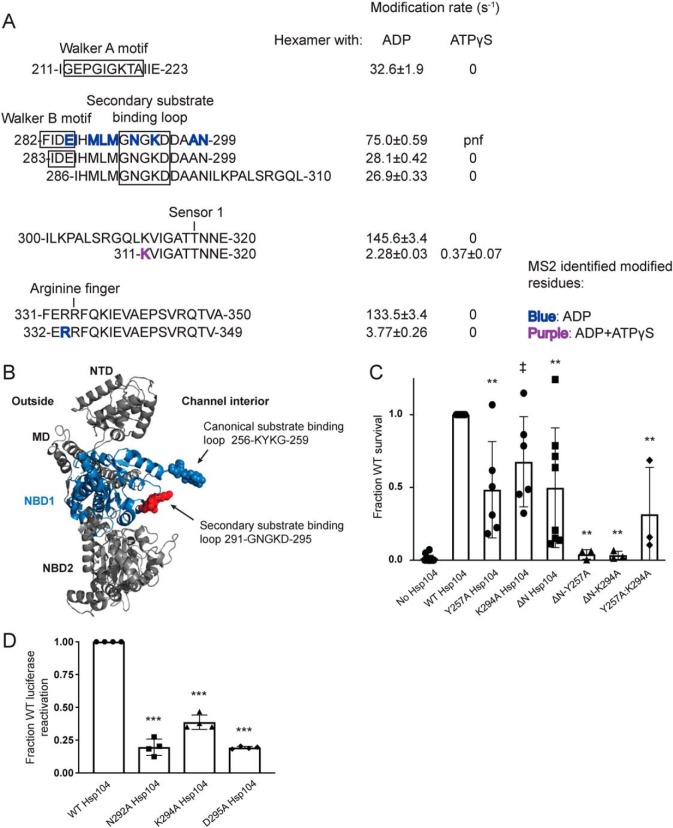
Figure 5.
Second substrate-binding loop in NBD1 is essential for Hsp104 disaggregase activity. A, peptide sequences and modification rates for regions of interest of Hsp104 NBD1. Residues shown in bold have been identified as modified by MS2 and are colored by the state in which they were found: blue, hexamer with ADP; purple, both hexameric states. B, NBD1 homology modeled on the tClpB crystal structure (PDB code 1qvr) shown in context of a rigid body fit Hsp104 monomer. NBD1 is shown in blue, with the canonical substrate-binding loop in blue spheres and the secondary substrate-binding loop in red spheres. C, after incubation at 37 °C for 30 min to induce Hsp104 expression, W303aΔhsp104 yeast carrying either empty vector or a plasmid encoding the indicated Hsp104 variant were heat-shocked for 20 min at 50 °C, immediately transferred to ice for 2 min, plated on SD-ura plates, and after a 2-day incubation at 30 °C, colonies were counted using an acolyte automated colony counter. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 3–14). Equal expression levels were confirmed by immunoblot. A one-way ANOVA with Dunnett's test was used to compare WT Hsp104 to the variants with ‡ denoting p = 0.05 and ** denoting p < 0.01. D, urea-denatured firefly luciferase aggregates were incubated with either WT or an Hsp104 substrate-binding loop 2 (291GNGKD295) variant, for 90 min at 25 °C in the presence of 2.6 mm ATP, 2.5 mm ATPγS, and an ATP-regenerating system (1 mm creatine phosphate and 0.25 μm creatine kinase). Reactivation of luciferase was then determined by measuring luminescence and converted to fraction WT activity for each condition. Data are displayed as scatterplot with bar representing mean ± S.D. (n = 3). A one-way ANOVA with Dunnett's test comparing variants to Hsp104-WT was performed with *** denoting p < 0.0001.