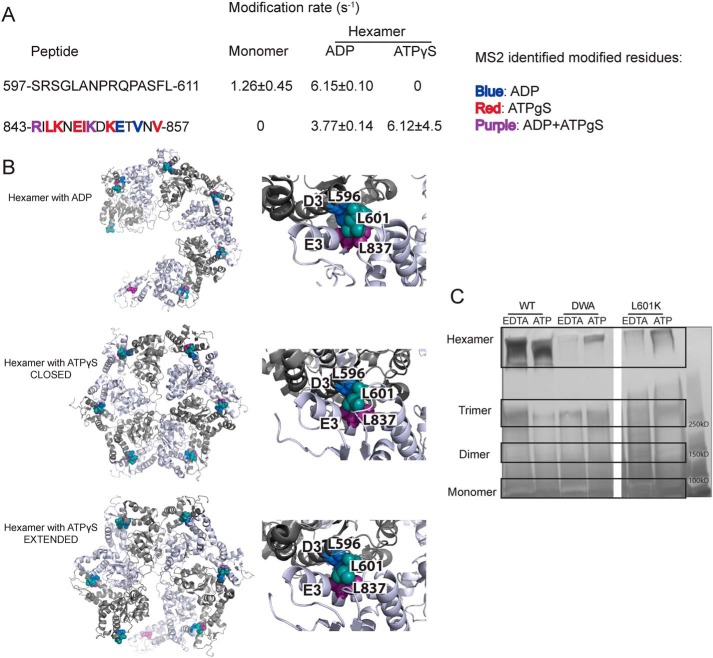
Figure 7.
Leu-601 plays an important role in Hsp104 hexamerization. A, peptide sequences and modification rates for regions of interest of Hsp104 NBD2. Residues shown in bold have been identified as modified by MS2 and are colored by the state in which they were found: blue, hexamer with ADP; red, hexamer with ATPγS; purple, both hexameric states. B, hexameric models (hexamer with ADP, PDB code 5vy8; hexamer with ATPγS closed, PDB code 5vjh, and extended, PDB code 5vya, conformations) of NBD2 generated from cryo-EM studies (16) showing the location of hydrophobic residues found on helices D3 and E3. On the right-hand side are higher magnification views of the interface between residues 586AIKAVSNAVRLSRSGL601 of the large subdomain of subunit 1 (helix D3) and residues 836ILNKLALRILKNEI849 of the small domain of subunit 2 (helix E3). Residues Leu-601 (large domain D3, teal), Leu-596 (large domain D3, blue), and Leu-837 (small domain E3, purple) are shown as spheres. C, glutaraldehyde cross-linking. Hsp104 shows a robust ability to form hexamers even in the presence of EDTA and absence of ATP. The double Walker A (DWA) mutant displays defects in hexamerization even in the presence of ATP. The mutant L601K, which resides in the proposed hexamer interface but outside of any secondary structure, also displays defects in the ability to form hexamers.