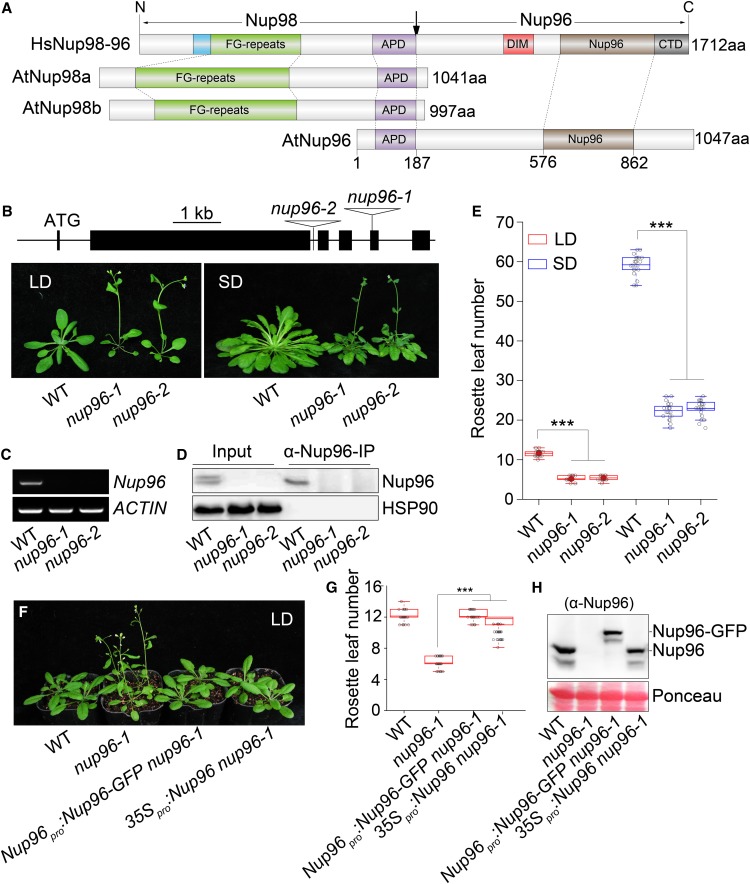
Figure 1.
Nup96 Acts as a Negative Regulator of Flowering in Arabidopsis.
(A) Domain structures of human Nup98-96 (HsNup98-96), Arabidopsis Nup98a and Npu98b (AtNup98a and AtNup98b), and Arabidopsis Nup96 (AtNup96). Domains are indicated as follows: the GLEBS motif (blue), the Phe-Gly repeat region (FG-repeats; green), the APD (purple), the domain invasion motif (DIM; red), the Nup96 domain (Nup96; brown), and the C-terminal domain (CTD; gray). The vertical arrow indicates the sites of autoproteolytic cleavage. aa, Amino acids.
(B) Scheme of Nup96 gene structure showing the positions of two T-DNA insertions (nup96-1 and nup96-2) and flowering phenotypes of the wild type (WT) and nup96-1 and nup96-2 mutants under long-day (LD) and short-day (SD) conditions. Black lines indicate introns. Black bars represent exons. Star codon ATG is indicated. Triangles refer to T-DNAs.
(C) RT-PCR analysis of Nup96 full-length transcripts in different genotypes. ACTIN was used as the control. WT, wild type.
(D) IP assay of Nup96 proteins in different genotypes. WT, wild type.
(E) Measurement of rosette leaf numbers of different genotypes (n > 20). Asterisks indicate significant differences according to Student’s t test (***, P < 0.001). WT, wild type.
(F) Flowering phenotypes of the wild type, nup96-1, and two complementation lines. WT, wild type.
(G) Measurement of rosette leaf numbers of genotypes shown in (F); n > 20. Asterisks indicate a significant difference according to Student’s t test (***, P < 0.001). WT, wild type.
(H) Immunoblot showing protein levels of endogenous Nup96 and Nup96-GFP in genotypes shown in (F) using anti-Nup96 antibodies. WT, wild type.