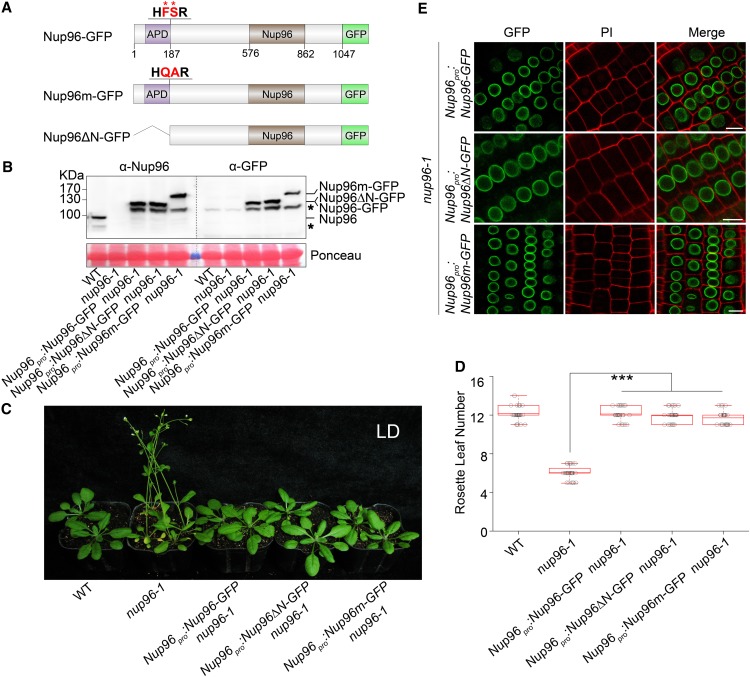
Figure 2.
Autoproteolytic Processing Is Not Required for the Physiological Activities of Nup96.
(A) Domain structures of Nup96-GFP, Nup96m-GFP, and Nup96ΔN-GFP fusion proteins. Domains of Nup96 are indicated as follows: the APD (purple box), the Nup96 domain (Nup96; brown box), and the rest of the regions (white box). The GFP fused to different versions of Nup96 are shown as a green box. The autoproteolytic cleavage site is shown as “HFSR,” and the key residues (F and S) used for mutations are labeled by asterisks and highlighted as red color. The numbers below the Nup96-GFP structure denote the positions in the amino acid sequence.
(B) Immunoblots showing the levels of endogenous Nup96, Nup96-GFP, Nup96m-GFP, and Nup96ΔN-GFP proteins in plants shown in (C) detected by anti-Nup96 and anti-GFP antibodies. Asterisks denote unknown bands. WT, wild type.
(C) Flowering phenotypes of the wild type, nup96-1, and transgenic lines expressing the proteins indicated in (A). WT, wild type.
(D) Rosette leaf numbers of genotypes shown in (C) at the time of flowering (n > 20). Box plots are used to display the distributions of data points (presented as circles). The lines in the box (from top to bottom) indicate the maximum, third quartile, mean, first quartile, and minimum. Asterisks indicate a significant difference according to Student’s t test (***, P < 0.001). WT, wild type.
(E) Subcellular distributions of Nup96-GFP, Nup96m-GFP, and Nup96ΔN-GFP fusion proteins indicated in (A) in plant root cells. Bars = 10 µm.