Figure 7.
Loss of Nup96 Function Promotes CO Accumulation, Resulting in an Early-Flowering Phenotype in Long Days.
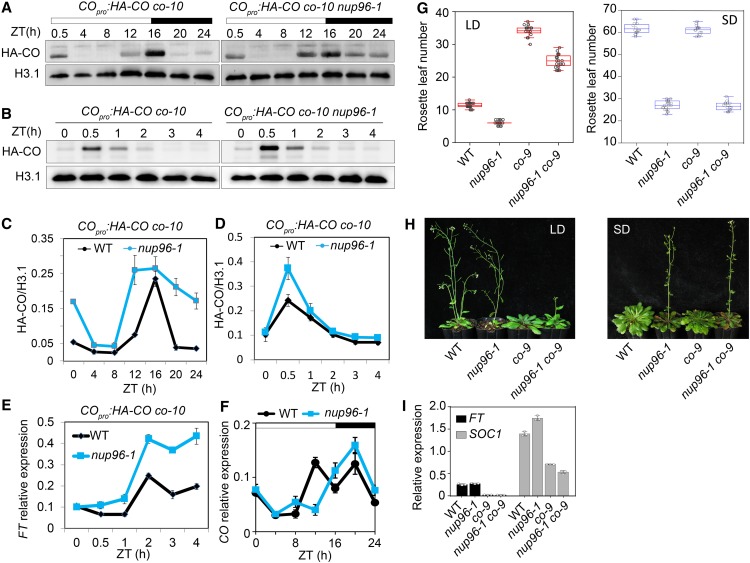
(A) and (B) CO protein levels in nuclear extracts from the indicated transgenic plants (COpro:HA-CO co-10 versus COpro:HA-CO co-10 nup96-1). Histone H3.1 (H3.1) was used as the loading control.
(C) and (D) Quantification of CO levels in immunoblots of (A) and (B), respectively. Values are means ± se (n = 3 biological repeats). WT, wild type.
(E) Expression levels of FT in the morning phase as in (B) and (D) in COpro:HA-CO co-10 versus COpro:HA-CO co-10 nup96-1 in long-day conditions for 9 d. Values are means ± se (n = 3 biological repeats). IPP2 (At3g02780) was used as a reference gene. WT, wild type.
(F) Diurnal expression of CO in the wild type (WT) and nup96-1 mutants grown in long-day conditions for 9 d. Values are means ± se (n = 3 biological repeats). TIP41 (At4g34270) was employed as a control gene.
(G) Rosette leaf numbers at the time of flowering for the genotypes of WT, nup96-1 and co-9 single mutants, and nup96-1 co-9 double mutants in long-day (LD) and short-day (SD) conditions (n > 20). Box plots are used to display the distributions of data points (presented as circles). The lines in the box (from top to bottom) indicate the maximum, third quartile, mean, first quartile, and minimum.
(H) Flowering phenotypes of WT, nup96-1 and co-9 single mutants, and nup96-1 co-9 double mutants grown in long days (left) and short days (right).
(I) Expression analysis of FT and SOC1 in different genotypes indicated in (H). Values are means ± sd (n = 3 biological repeats). WT, wild type.