Figure 7.
Overexpression of ALT TAS3 Enhances Nodule Formation.
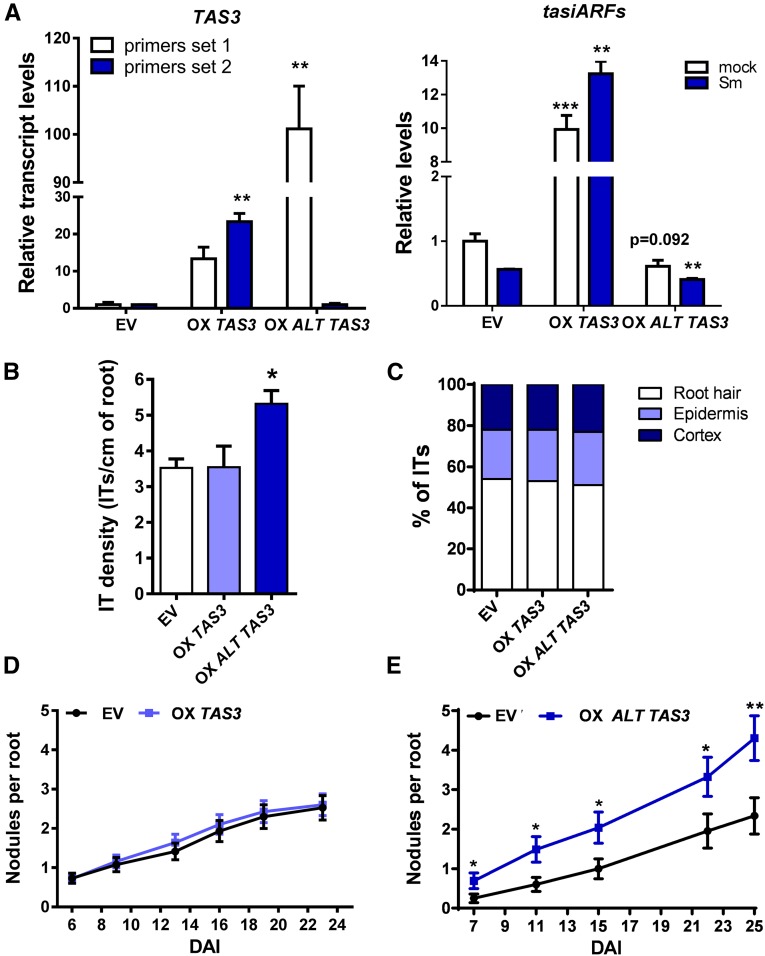
(A) Relative transcript levels of TAS3 transcripts using the set of primers 1 and 2 (red and black arrows in Figure 6A, respectively) measured in roots transformed with the EV or with constructs to overexpress TAS3 (OX TAS3) or ALT TAS3 (OX ALT TAS3). Expression levels were determined by RT-qPCR, normalized to HIS3L, and expressed relative to the EV sample. Expression levels of tasiARFs in EV, OX TAS3, or OX ALT TAS3 roots were determined by stem-loop RT-qPCR, normalized to U6 RNA, and expressed relative to the EV sample. Each bar represents the mean ± se of three biological replicates (whole root tissue from independent experiments performed in different days), with three technical replicates each. Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01, ***P < 0.001) in an unpaired two-tailed Student’s t test.
(B) Infection threads (ITs) per centimeter of root developed at 7 DAI in EV, OX TAS3, and OX ALT TAS3 roots. The asterisk indicates statistically significant differences compared with EV with P < 0.05 in an unpaired two-tailed Student’s t test.
(C) Progression of infection events in EV, OX TAS3, and OX ALT TAS3 roots. Infection events were classified as ITs that end in the root hair, in the epidermal cell layer, or reach the cortex at 7 DAI.
(D) Nodules per root formed in EV and OX TAS3 roots at indicated time points after inoculation with S. meliloti.
(E) Nodules per root formed in EV and OX ALT TAS3 roots at time points after inoculation with S. meliloti. Asterisks indicate statistically significant differences as compared with EV (*P < 0.05, **P < 0.01) in an unpaired two-tailed Student’s t test (Supplemental Data Set 11). Error bars in (B) to (E) represent the mean ± se of three biological replicates (independent experiments performed in different days) each with at least 50 roots.