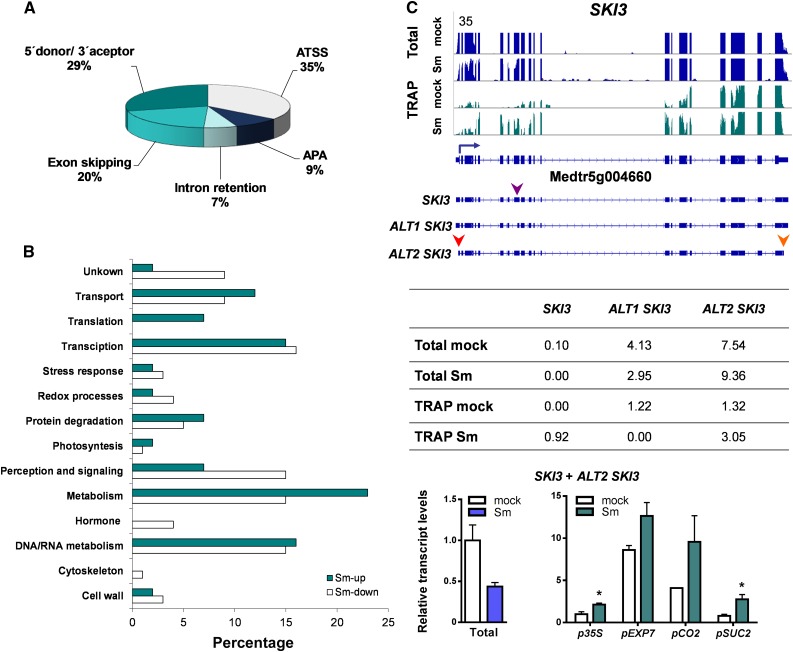
Figure 8.
Differential Translation of Alternative Transcript Variants.
(A) Pie chart classification of translationally regulated transcript variants produced by ATSS, APA, or alternative splicing, including intron retention, exon skipping, and alternative 5′ donor or 3′ acceptor sites.
(B) Functional classification of genes with translationally regulated alternative transcript variants based on GO and manual inspection of individual genes. The percentage of DETs are presented.
(C) Expression of alternative transcript variants of SKI3. (Top) Transcript isoforms of SKI3 and read coverage in total (blue) and TRAP (green) samples of roots inoculated with S. meliloti (Sm) or with water (mock). SKI3 gene model and its transcript variants are illustrated along the x axis. The blue arrow indicates the canonical translation initiation codon and the arrowheads indicate an alternative 3′ acceptor site (purple), an ATSS (red), and an APA (orange). (Middle) Reads corresponding to the different transcript variants of SKI3 in total and TRAP samples expressed as FPKM. (Bottom) RT-qPCR validation and tissue-specific analysis of selected transcript variants. Abundance of specific transcript variants of SKI3 in total RNA samples or in TRAP RNA samples of nearly all cells (p35S), epidermal (pEXP7), cortical (pCO2), or phloem companion (pSUC2) cells from roots inoculated with S. meliloti (Sm) or with water (mock). Total RNA samples were obtained from p35S:FLAG-RPL18 roots. Expression values were determined by RT-qPCR, normalized to HIS3L, and expressed relative to the mock total sample or the mock p35S sample. Each bar represents the mean ± se of two independent biological replicates, with three technical replicates each. An asterisk indicates statistically significant differences in Sm compared with mock samples (*P ≤ 0.05) in an unpaired two-tailed Student’s t test (Supplemental Data Set 11).