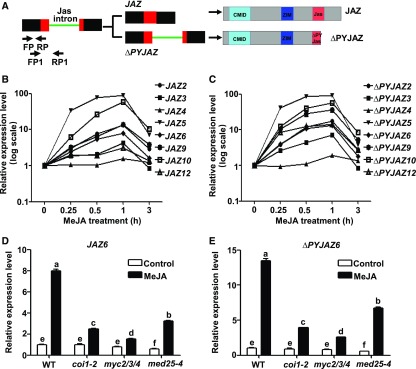
Figure 1.
MeJA-Induced Generation of ∆PYJAZs Depends on MYC2 and MED25.
(A) Schematic diagram of Jas intron-dependent AS of JAZ and its corresponding translational products. In the gene structure, boxes represent exons, lines represent introns, red boxes represent Jas motif-encoding regions, and the green line represents the Jas intron. Arrows indicate forward primers (FP or FP1) and reverse primers (RP or RP1) used for RT-qPCR to amplify transcripts in which the Jas intron is spliced (FP/RP) or retained (FP1/RP1). In the protein structures of JAZ and ∆PYJAZ, cyan boxes represent CMID, blue boxes represent the ZIM domain, and red boxes represent the Jas motif.
(B) and (C) RT-qPCR showing fully spliced transcripts (B) or Jas intron-retained transcripts (C) of Jas intron-contained JAZ genes in MeJA-treated wild-type seedlings. Ten-d–old seedlings were treated without or with 100 μM of MeJA for the indicated time before RNA extraction.
(D) and (E) RT-qPCR showing JAZ6 (D) and ΔPYJAZ6 (E) expression in response to MeJA in wild type, coi1-2, myc2/3/4, and med25-4 seedlings. Ten-d–old seedlings were treated with 100 μM of MeJA for 60 min before RNA extraction.
(B) to (E) Expression levels of target genes were normalized to ACTIN7, and the expression levels in wild type without MeJA treatment were arbitrarily set to 1. Data shown are mean values of three biological repeats with sd.
(D) and (E) Statistical analysis was performed via one-way ANOVA (Supplemental File); bars with different letters are significantly different from each other (P < 0.01).