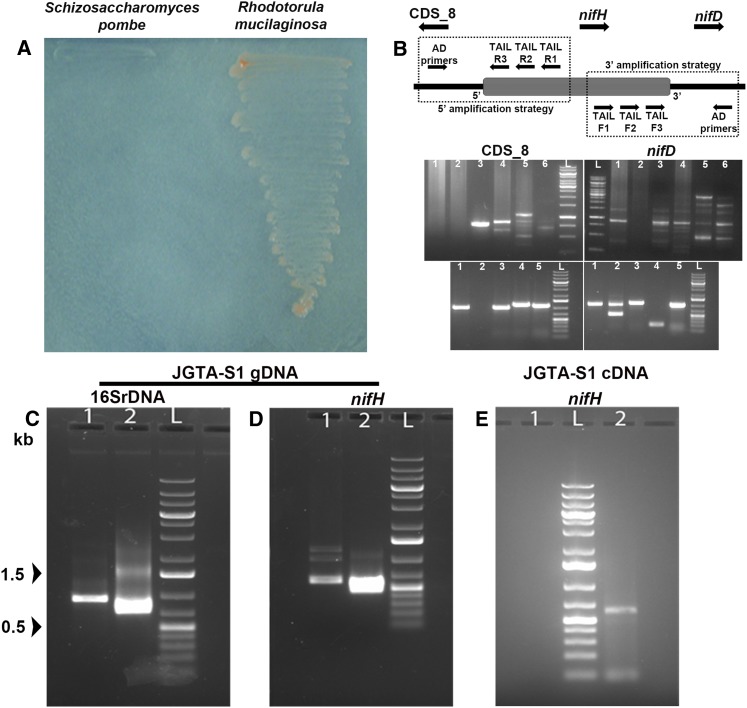
Figure 5.
R. mucilaginosa JGTA-S1 Contains Bacteria Related to P. stutzeri as an Endosymbiont.
(A) JGTA-S1 was streaked onto N-free medium along with a laboratory strain of yeast S. pombe D18 h− to examine its diazotrophic nature.
(B) Flanking genes of P. stutzeri nifH from JGTA-S1 genomic DNA amplified using TAIL-PCR. The directions of the genes are represented as in P. stutzeri DSM4166. (Top, left) Amplification of the CDS_8-homolog. (Top, right) Amplification of the nifD-homolog using TAIL R3 or TAIL F3 gene-specific primers together with different AD primers. Lane 1, AD1; lane 2, AD2; lane 3, AD3; lane 4, AD4; lane 5, AD5; lane 6, AD6; lane L, DNA markers. (Bottom) Representative amplicons after colony PCR from putative CDS_8 and nifD clones.
(C) and (D) P. stutzeri 16SrDNA and nifH gene–specific primers were used to amplify P. stutzeri 16SrDNA (C) and nifH (D) from JGTA-S1 genomic DNA. (C) Lane 1, primary PCR product; lane 2, nested PCR with internal primers. (D) Lane 1, primary PCR product; lane 2, nested PCR with internal primer.
(E) RNA was isolated from JGTA-S1 and the cDNA used to amplify P. stutzeri–specific nifH. Lane 1, no DNA control; lane 2, nifH gene amplicon using nifH internal primers; lane L, DNA markers.