Figure 2. Engineering a Genomically Recoded E. coli Strain for T7RNAP Overexpression.
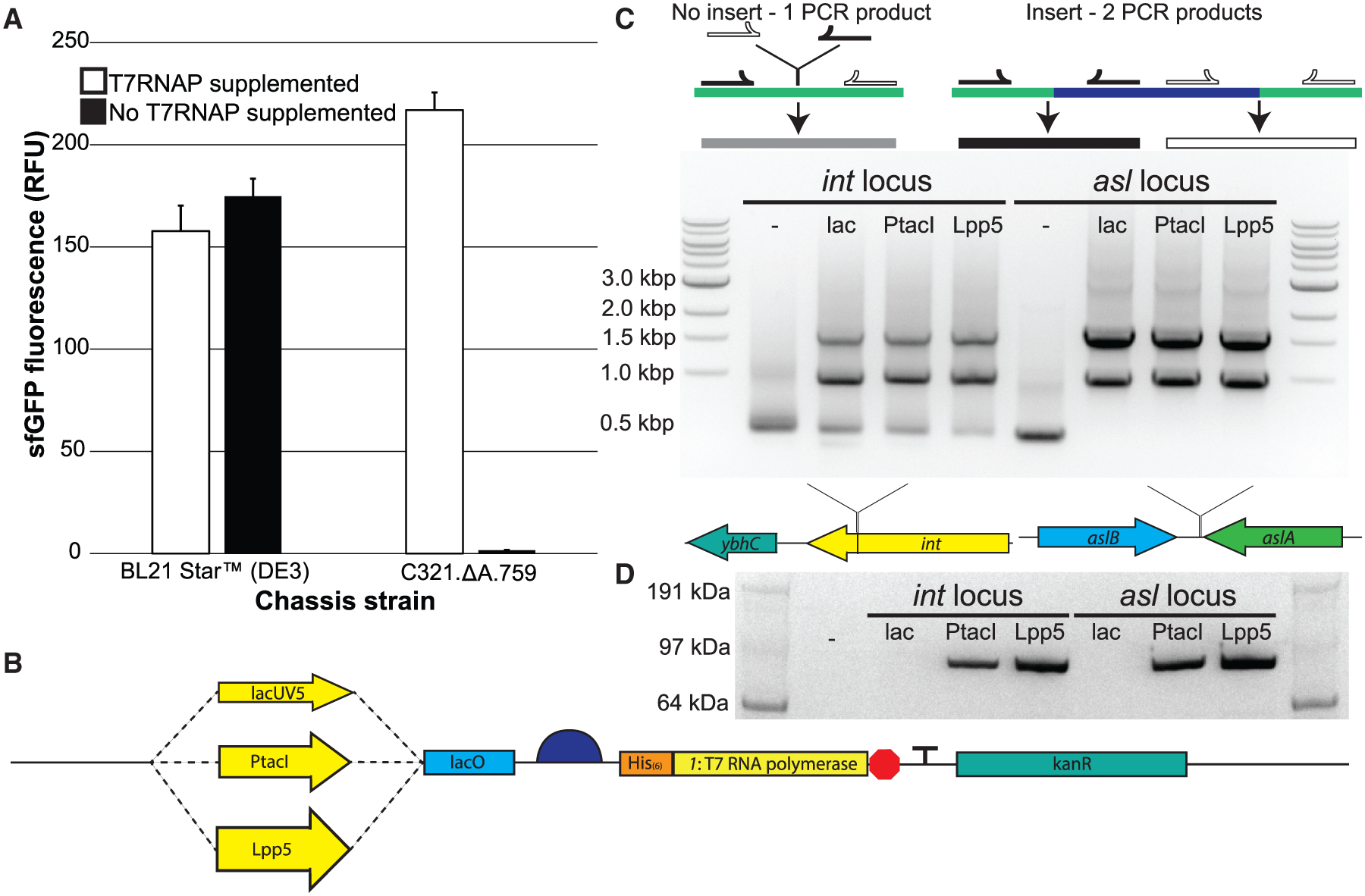
(A) sfGFP fluorescence in vitro from cell extracts derived from induced BL21 Star (DE3) cells as well as C321.ΔA.759 cells, both with and without supplementation with purified T7RNAP.
(B) Schematic of the synthetic genomic insert used in this study to introduce the gene encoding the T7RNAP into the genome of C321.ΔA.759.
(C) Top: diagram illustrating the PCR-based detection scheme for successful genomic integration of the synthetic T7RNAP cassette into strain C321.ΔA.759. Middle: shown are the MASC PCR products generated from C321.ΔA.759 and the six T7RNAP-expressing strains generated in this study, run on an agarose gel. The dash indicates the unaltered strain without any insert incorporated. Bottom: depiction of the two insertion loci used.
(D) α-His western blot analysis of protein samples derived from IPTG-induced populations of C321.ΔA.759 and the six T7RNAP-expressing strains generated in this study. The His-tagged T7RNAP version used has a molecular weight of ~100 kDa. The dash indicates the unaltered strain without any insert incorporated.
