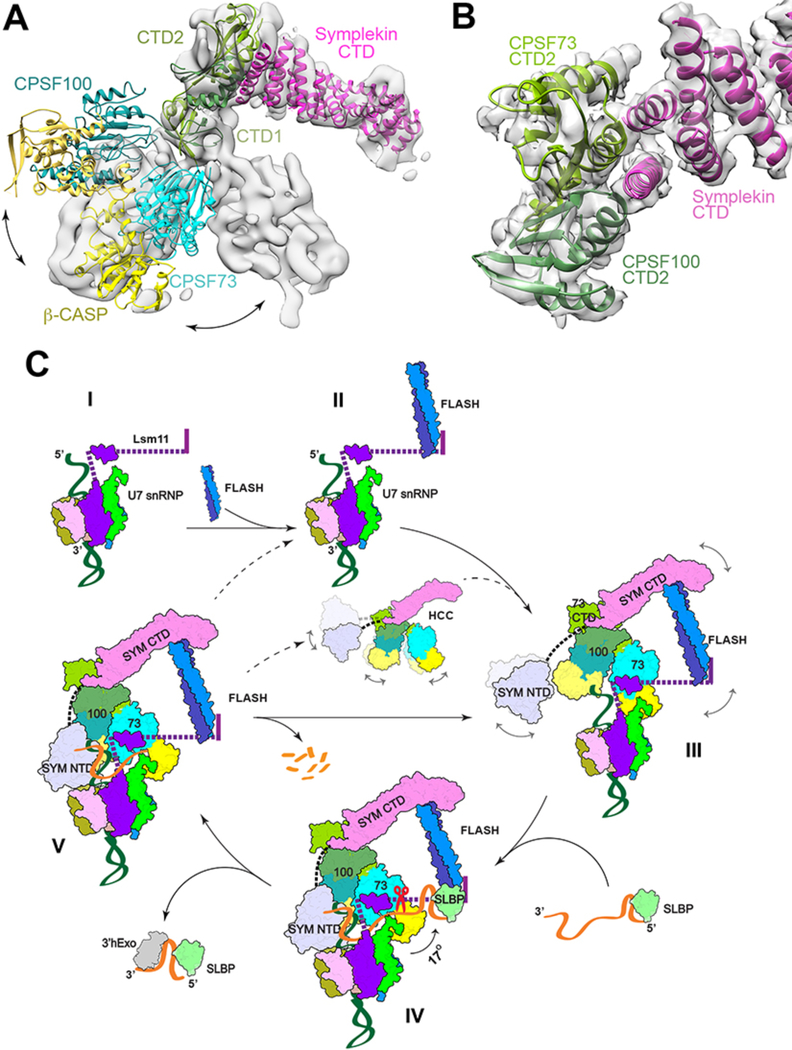
Figure 4. Schematic of histone pre-mRNA 3′-end processing cycle.
(A) Significant structural differences of HCC in an active state compared to an inactive state. The structure of HCC observed here is docked into the EM density for mCF (gray surface) (25), using the symplekin CTD as the reference. (B) Schematic drawing of the CTD2 domain complex of CPSF73 (light green) and CPSF100 (darker green) and the N-terminal segment of the symplekin CTD (magenta). The CTD complex of IntS9 and IntS11 (28) was docked into the EM density at 4.1 Å resolution (transparent surface) using Chimera. Panels A and B produced with Chimera. (C) A putative model for histone pre-mRNA 3′-end processing cycle. The machinery is assembled from the U7 snRNP (state I) with the recruitment of FLASH (II) and HCC (III), followed by the recognition of the pre-mRNA for CPSF73 and HCC activation and pre-mRNA cleavage (IV). The machinery is likely highly dynamic before the binding of the authentic pre-mRNA, and the possible flexible regions are indicated with the curved arrows and dashed lines. After the cleavage (V), the downstream product is degraded by an exonuclease activity and the machinery can be recycled directly (solid arrow), or possibly disassembled followed by re-assembly. State IV corresponds to the structure reported here, with the scissors indicating cleavage by CPSF73, and the other states are models.