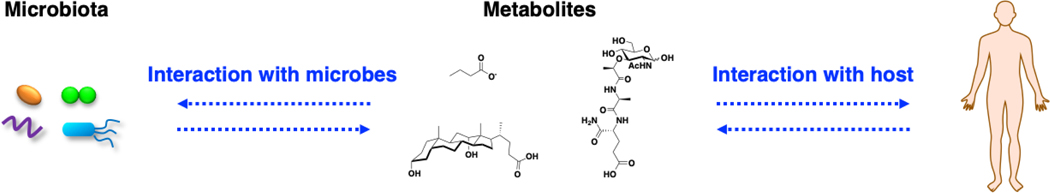
Graphical Abstract
The advances in bioorthogonal chemical reporters have afforded new tools to explore the targets and functions of specific metabolites in diverse microbes. Here we summarize the development and application of metabolite chemical reporters to study fundamental pathways in bacteria as well as microbiota mechanisms in health and disease.
Frontispiece
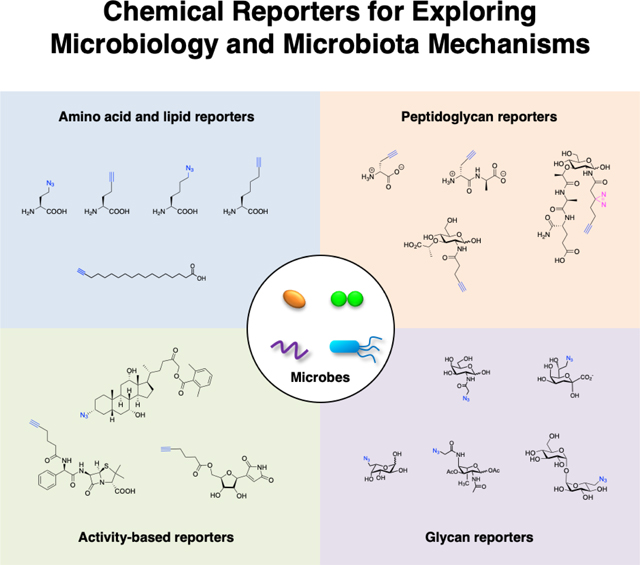
Keywords: Bioorthogonal chemistry, Chemical biology, Chemical reporter, Microbiology, Microbiota
INTRODUCTION
Microbes mediate fundamental processes of life on the planet and interact directly with humans to regulate health and disease. The interactions between microbes with the environment and their hosts are often mediated by small molecules or metabolites (Fig. 1). Understanding the chemical dialogue of microbes is therefore crucial for maintaining healthy ecosystems and modulating their interactions with plants, insects, animals and humans. Given the remarkable diversity of microbes on the planet and their unique metabolic pathways, they encode an enormous capacity to generate unique small molecules that are important for microbial ecology and interaction with their specific hosts. New chemical tools are therefore continually needed to investigate the functions of specific microbial metabolites and biosynthetic pathways for individual microbial species as well as complex microbial communities. Herein, this review will survey bioorthogonal chemical reporters of microbial metabolites that have been developed to investigate fundamental microbial pathways (translational, cell wall, glycan and lipid biosynthesis as well as microbial communication) and then review the applications of these chemical reporters to explore microbiota mechanisms.
Figure 1.
Metabolites from host metabolism or the environment (diet and microbiota) can interact with microbes and the host to regulate physiology and disease. Representative metabolites: butyrate, deoxycholic acid and muramyl dipeptide.
CHEMICAL REPORTERS FOR MICROBIOLOGY
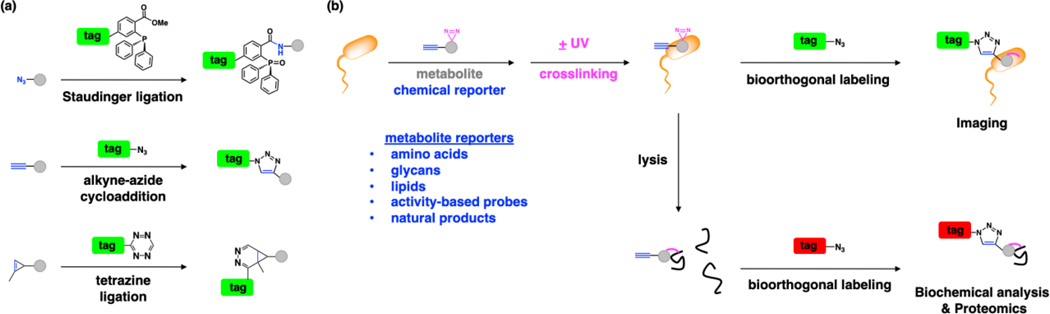
The advances in chemical biology have provided new tools to explore microbiology and microbiota mechanisms. Notably, the development of bioorthogonal chemical reactions (Staudinger ligation [1], azide-alkyne cycloadditions [2, 3] and tetrazine ligations [4]) has provided new approaches to label biomolecules and study their functions in vitro, in cells and whole animals (Fig. 2a) [5–7]. In particular, the use of relatively small and uniquely reactive functional groups such as azides, alkynes and activated alkenes has enabled the modification of metabolites to visualize their direct interaction with diverse biomolecules (Fig. 2b) [5, 8]. These “chemical reporters” provide more sensitive alternatives to classical radioactive tracers and enable the imaging and biochemical labeling of metabolite-targets (Fig. 2b). Chemical reporters of diverse metabolites, amino acids, lipids, glycans, synthetic probes, and natural products, have now been generated by many research groups to study metabolite-interactions and trafficking in eukaryotic cells and animals [5, 9–13]. In this review, we focus on the development and application of chemical reporters to study fundamental pathways in bacteria as well as exploration of microbiota mechanisms in health and disease.
Figure 2.
Chemical strategy for exploring metabolite trafficking and protein interactions in bacteria. (a) Representative bioorthogonal labeling reactions; Staudinger ligation [1], click chemistry (copper(I)-catalyzed azide-alkyne cycloaddition) [2, 3] and tetrazine ligation [4], enable selective labeling of azide, alkyne and activated-alkene modified metabolites, respectively. (b) These advances in bioorthogonal labeling have inspired the development of diverse azide, alkyne and activated-alkene functionalized metabolites as chemical reporters. Chemical reporter-labeled targets can then be reacted with complementary-functionalized fluorescent dye or affinity tag for imaging and/or proteomic applications. For non-covalent metabolite-protein interactions, chemical reporters can be further modified with diazirines for photocrosslinking.
Amino acid reporters
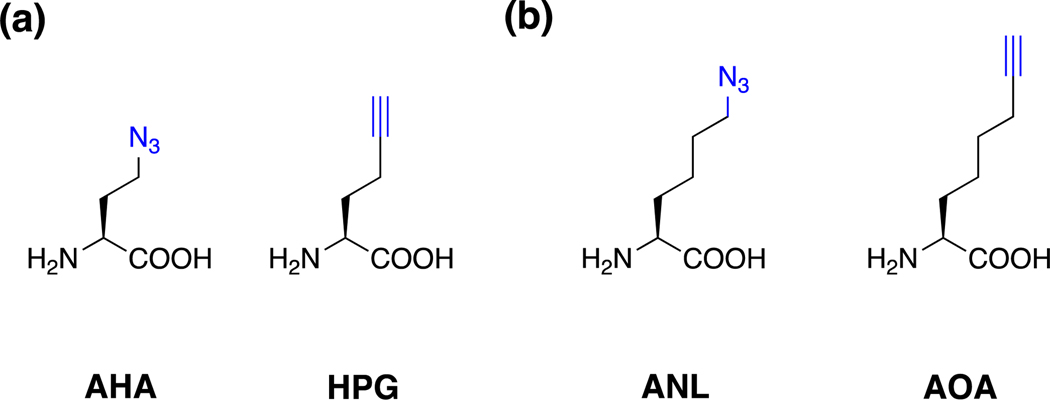
The first chemical reporter for labeling bacteria was a methionine (Met) analog. Following the development of the Staudinger ligation by the Bertozzi group (Fig. 2a) [1], an azide derivative of Met (azido-homoalanine, AHA) was synthesized and demonstrated to label bacterial proteins (Fig. 3a) [9]. This allowed the use of AHA and an alkyne-analog, homopropargylalanine (HPG) [9, 14] to function as chemical reporters of protein synthesis by the Tirrell laboratory (Fig. 3a) [15]. After these pioneering studies demonstrated that azide- and alkyne-amino acids could be incorporated into bacteria, additional chemical reporters (Fig. 3b) and orthogonal tRNA synthetase mutants were developed for selective monitoring of bacterial protein translation for intra- and inter-species interactions [16] as well as infection of mammalian cells [17, 18]. For example, the Tirrell and Newman laboratories have subsequently utilized orthogonal amino acid reporter labeling, also termed “bioorthogonal non-canonical amino-acid tagging (BONCAT), for selective proteomic analysis of Pseudomonas aeruginosa subpopulations, which revealed differential protein expression and antibiotic tolerance in biofilms [19]. Alternatively, our laboratory has employed selective amino acid reporter labeling in Salmonella to characterize the bacterial proteome during replication in macrophages [18]. The studies highlight the utility of amino acid reporters for time-resolved and cell-selective analysis of bacterial translation in different cellular states and multicellular interactions.
Figure 3.
Amino acid chemical reporters for monitoring protein synthesis. (a) AHA and HPG [9, 14]. (b) Orthogonal amino acid reporters for bacteria-specific labeling using mutant tRNA synthetases [17, 18].
Peptidoglycan reporters
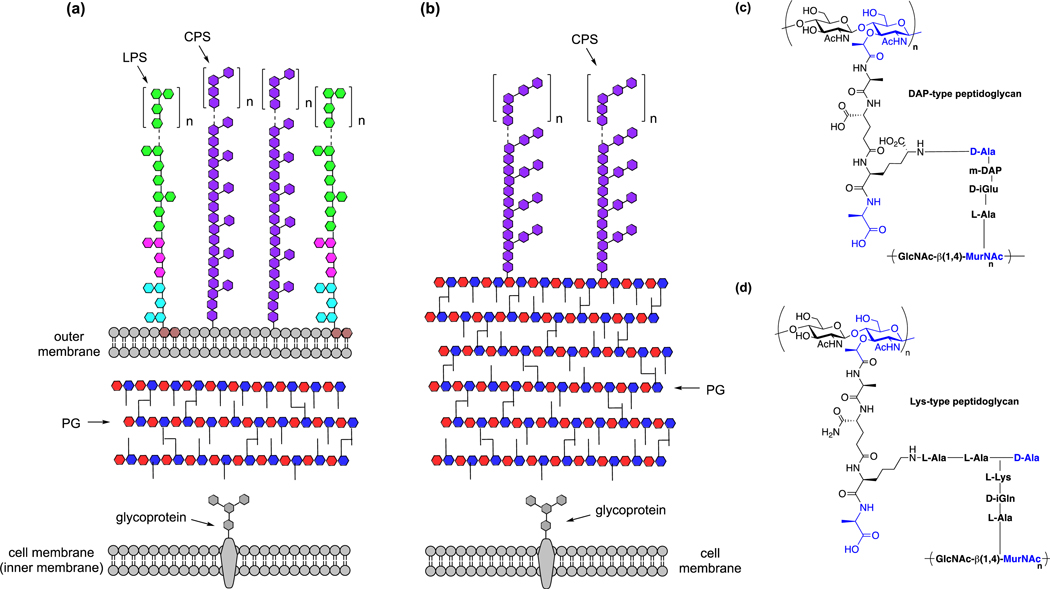
In addition to amino acid reporters for bacterial translation, chemical reporters have been developed to monitor peptidoglycan biosynthesis. Peptidoglycan is a crosslinked glycopeptide matrix that functions as the cell wall or exoskeleton of bacteria, which dictates their shape and tolerance to environmental stress (Fig. 4a) [20]. Gram-negative bacteria have an outer membrane and thinner peptidoglycan (~5 nm) [21] (Fig. 4a), whereas Gram-positive bacteria contain thicker multilayered peptidoglycan (~20–50 nm) and only one cell membrane (Fig. 4b). The repeating units of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) serve as the glycan backbone of peptidoglycan, which is then further crosslinked by branched peptides. The peptide crosslinking varies between bacterial species and can be classified as diaminopimelic acid (DAP)- or lysine (Lys)-type, depending on the structure of stem amino acids (Fig. 4c,d).
Figure 4.
Bacterial cell surface architecture and peptidoglycan composition. (a) Schematic illustration of cell membranes and peptidoglycan of Gram-negative bacteria. LPS, lipopolysaccharides; CPS, capsular polysaccharides; PG, peptidoglycan. (b) Schematic illustration of cell membrane and peptidoglycan of Gram-positive bacteria. CPS, capsular polysaccharides; PG, peptidoglycan. (c) Structure of DAP-type peptidoglycan. (d) Structure of Lys-type peptidoglycan.
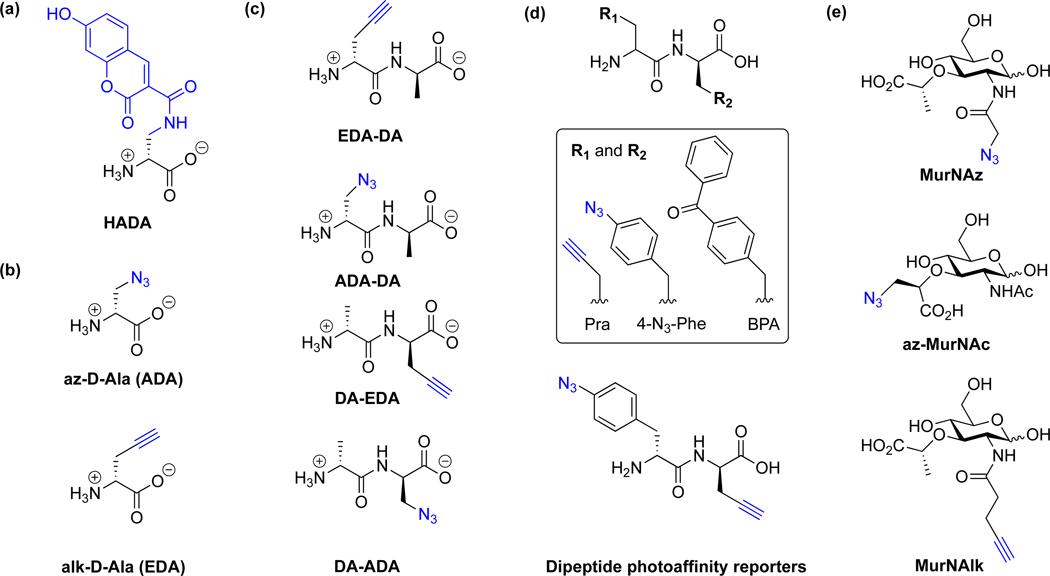
Peptidoglycan synthesis, editing, and turnover are highly dynamic and regulated during cell division and in response to environmental stress. The underlying mechanisms of peptidoglycan biosynthesis and turnover in different bacteria species are still not completely understood and have been further elucidated by the development of specific chemical reporters. Notably, the first set of fluorescein-MurNAc pentapeptide reporters by Nishimura laboratory enabled fluorescent labeling of both Gram-negative and Gram-positive bacterial cell wall [22]. Moreover, fluorescent-D-Ala analogs from the VanNieuwenhze laboratory have revealed temporal regulation of peptidoglycan synthesis in diverse bacteria (Fig. 5a) [23] and helped demonstrating the existence of peptidoglycan in Chlamydia [24], which only replicates inside host cells and was previously challenging to characterize. Alkyne-, azide- and cyclooctyne-D-Ala reporters from the Bertozzi laboratory have also facilitated the analysis of peptidoglycan turnover in bacteria (Fig. 5b) [25, 26]. For example, alk-D-Ala has been recently used by the Galan laboratory in Salmonella Typhi to evaluate peptidoglycan editing by a specific L,D-transpeptidase that controls the muramidase-dependent secretion of typhoid toxin [27]. Since bacteria can recycle more than D-Ala, dipeptide reporters have also been developed to monitor the de novo biosynthesis and turnover of peptidoglycan (Fig. 5c) [28, 29]. In addition, dipeptide reporters have also been employed to monitor L,D-transpeptidase-mediated peptidoglycan cross-linking [30] as well as photocrosslinking of lipid II-binding proteins (Fig. 5d) [31].
Figure 5.
Chemical reporters for metabolic labeling of peptidoglycan. (a) D-Ala fluorophore analog (HADA) [23]. (b) D-Ala reporters [25]. (c) Dipeptide reporters [28, 29]. (d) Dipeptide photoaffinity reporters [31]. (e) MurNAc reporters [32].
In addition to amino acids and peptides, reporters of MurNAc have also been developed to monitor the biosynthesis and turnover of the bacterial cell wall glycan backbone (Fig. 5e). For E. coli and related enterobacteria as well as most Gram-positive bacteria, peptidoglycan turnover is carried out by MurQ pathway, in which MurNAc is transformed via GlcNAc-6P and GlcN-6P before transferred to peptidoglycan synthetic pathway. Alternatively, peptidoglycan turnover in many Gram-negative bacteria is carried out by the MurU pathway, in which MurNAc is converted to UDP-MurNAc for peptidoglycan synthesis via phosphorylation and dephosphorylation reactions [33–35]. With this in mind, the Grimes laboratory synthesized azide- and alkyne-reporters (Fig. 5e) and demonstrated that they can be incorporated into bacteria that contain AmgK and MurU enzymes for MurNAc recycling [32]. For bacteria without MurNAc recycling enzymes, UDP-MurNAc reporters can be used to bypass this pathway and label peptidoglycan for subsequent imaging studies [36]. Alternatively, N-acetyl cysteamine reporters can also be used to label and visualize peptidoglycan acylation [37]. Given that peptidoglycan biosynthetic pathways differ between different bacterial species, these D-amino acid, MurNAc and other chemical reporters should provide excellent tools to experimentally identify bacteria and specific enzymes involved peptidoglycan editing and recycling.
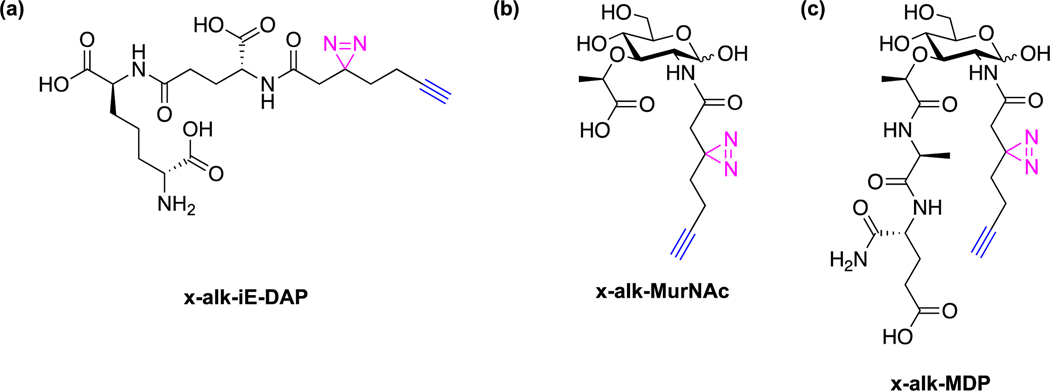
To explore peptidoglycan metabolite protein targets, our laboratory has synthesized photoaffinity reporters of key bacterial cell wall metabolites. In particular, we synthesized the alkyne- and diazirine-functionalized photoaffinity reporters ofγ-d-glutamyl-meso-diaminopimelic acid (iE-DAP), MurNAc and muramyl-dipeptide (MDP) (Fig. 6) and demonstrated that they can directly crosslink their respective cognate immune receptors, NOD1 and NOD2, in mammalian cells [38]. Interestingly, the MDP reporter also crosslinked Arf GTPases and induced the formation of GTP-Arf6:NOD2 complex in mammalian cells, which was abrogated with loss-of-function NOD2 mutants associated with Crohn’s Disease [38]. Further development and application of these peptidoglycan metabolite reporters should reveal additional cellular factors that may be important for host sensing of microbes and innate immunity.
Figure 6.
Peptidoglycan metabolite photoaffinity reporters. (a) iE-DAP photoaffinity reporter [38]. (b) MurNAc photoaffinity reporter [38]. (c) MDP photoaffinity reporter [38].
Bacterial glycan reporters
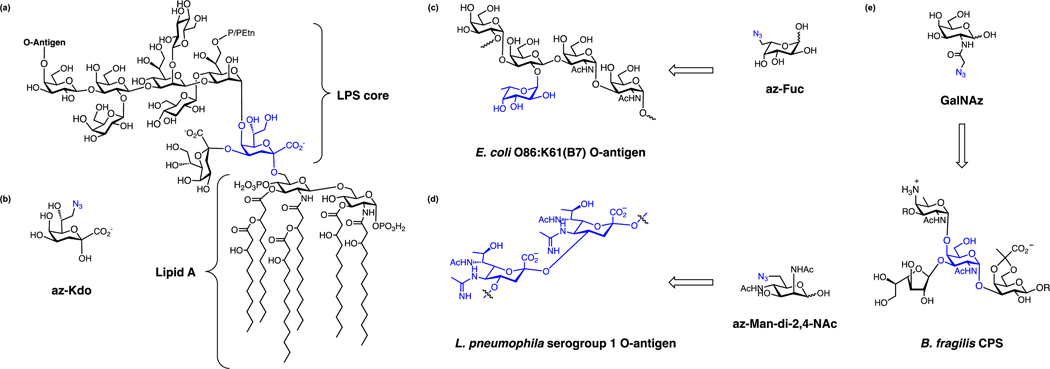
Chemical reporters have also been developed for glycans that decorate the cell surface of bacteria and post-translationally modify proteins. For example, glycan reporters have been developed for labeling lipopolysaccharides (LPS) (Fig. 7a), which are key glycolipids that helps maintain outer-membrane permeability and important for host-bacteria recognition [39]. LPS is composed of lipid A, inner core oligosaccharide, outer core oligosaccharide and a long-chain O-antigenic polysaccharide (O antigen) (Fig. 7a). The structures of lipid A and inner core oligosaccharide are relatively conserved among different bacteria although some species can modify these structures of LPS. Notably, 3-deoxy-D-manno-oct-2-ulosonic acid (Kdo) is a glycan that is selectively present in inner core of LPS and can be labeled by an azido-Kdo reporter (az-Kdo) (Fig. 7b) [40]. This az-Kdo reporter developed by the Vauzeilles laboratory labels Gram-negative bacteria such as E. coli, S. Typhimurium and Legionella pneumophila [40, 41], but does not label Gram-positive bacteria like B. subtilis or S. aureus, nor Shewanella oneidensis, which uses 8-amino-8-deoxy-Kdo instead of Kdo [40, 41].
Figure 7.
Chemical reporters for metabolic labeling of bacterial surface glycans. (a) Structure of LPS core and lipid A. (b) az-Kdo [40]. (c) az-Fuc and its target, E. coli O-antigen [42]. (d) az-Man-di-2,4-NAc and its target, L. pneumophila serogroup 1 O-antigen [43]. (e) GalNAz and its labeling of CPS [44].
In contrast to the inner core, O antigen outer glycans of LPS are more structurally diverse and highly variable. For example, there are more than 180 O antigens in E. coli and 46 O antigens in Salmonella strains [45, 46]. To label the O-antigen of E. coli O86, a fucose reporter was developed in conjunction with genetic engineering developed by the Wang laboratory [42]. An engineered E. coli O86 strain, E. coli O86:B7Δgmd-fcl(fkp), has been deleted of GDP-mannose dehydratase (Gmd) and GDP-fucose synthetase (Fcl) so that the bacteria cannot synthesize GDP-fucose de novo. A plasmid expressing a bifunctional enzyme possessing both fucokinase and pyrophosphorylase activity (FKP) is introduced so that the bacteria can utilize fucose to make GDP-fucose. An azido-fucose (az-Fuc) reporter can thus label the surfaces of the engineered E. coli O86:B7Δgmd-fcl(fkp) (Fig. 7c) [42]. Alternatively, L. pneumophila, which is a pathogenic bacterium, presents a variety of 16 serogroups [47]. The O-antigen of L. pneumophila serogroup 1 contains 5-N-acetimidoyl-7-N-acetyl-legionaminic acid (Leg), while serogroup 3, 4, 5, 6 contain an isomer of Leg, namely 5-N-acetimidoyl-7-N-acetyl-4-epi-legionaminic acid (4eLeg) [48]. The Vauzeilles laboratory also generated an azide derivative of Leg precursor (6-azido-2,4-deoxy-Man-diNAc) (Fig. 7d) and showed that it could label L. pneumophila serogroup 1, 3, 4, 5, 6 but not serogroup 7 or other species of Legionella, E. coli and P. aeruginosa [43].
In addition to LPS, many bacteria have capsular polysaccharides (CPS) on their surfaces (Fig. 4a,b). CPS are high-molecular-weight polymers with repeat units of monosaccharides such as GlcNAc or N-acetylgalactosamine (GalNAc), which can shield the bacteria from the host immune system by preventing either phagocytosis or complement-mediated lysis [49, 50]. CPS may also function as microbe-associate molecular patterns (MAMPs) and prime the immune system for tolerance towards commensal bacteria [51]. The compositions of CPS vary in different bacteria. For example, 80 capsular serotypes have been reported for E. coli, and 93 capsular serotypes have been reported for Streptococcus pneumoniae [49, 50, 52, 53]. To label CPS, the Kasper laboratory showed that an azide derivative of GalNAc (GalNAz) (Fig. 7e), can be used for metabolic labeling and fluorescence imaging of Gram-negative gut anaerobes such as Bacteroides fragilis, B. ovatus, and B. vulgatus [44]. These studies demonstrate that chemical reporters can label bacterial glycans and target specific LPS/CPS positions and serotypes depending on the expression and promiscuity of biosynthetic pathways.
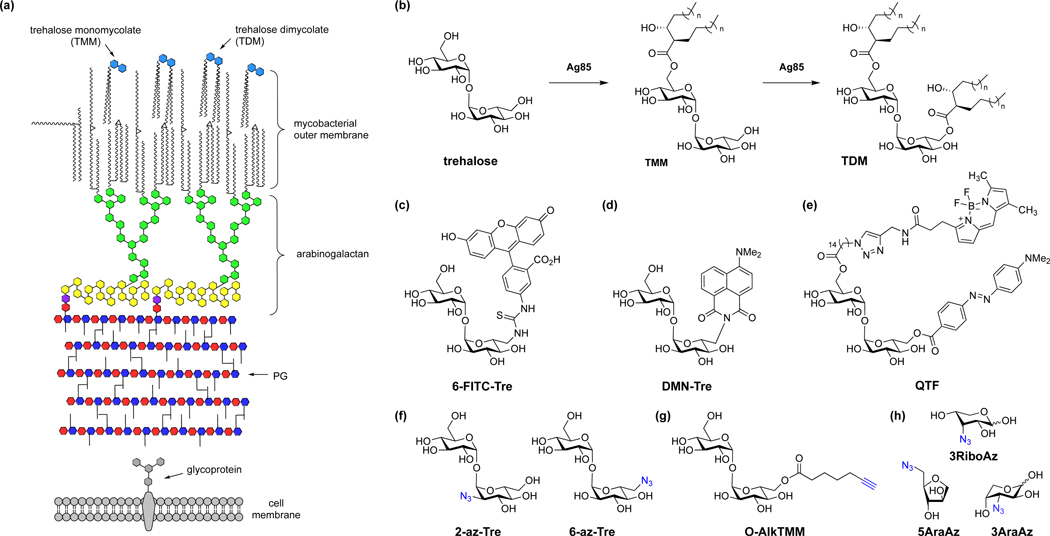
In contrast to other bacteria, mycobacteria has a unique cell envelope that contains peptidoglycan connected to a highly branched arabinogalactan and lipid-rich membrane of long-chain mycolic acids as well as trehalose glycolipids (Fig. 8a) [54]. The most abundant glycolipids in the mycobacterial outer membrane are trehalose monomycolate (TMM) and trehalose dimycolate (TDM) (Fig. 8b). To visualize mycobacteria and monitor biosynthesis of its cell envelope, several reporters for trehalose and its derivatives have been developed by the Davis [55], Bertozzi [56], Swarts [57] and Kiessling [8] laboratories. For example, trehalose analogs have been developed for visualization of mycobacteria cell division (Fig. 8e) [8], visualization of antibiotic activity on mycobacterial cell wall (Fig. 8c) [58], and rapid detection of live Mycobacterium tuberculosis (Mtb) in sputum using solvatochromic dyes for clinical TB diagnosis (Fig. 8d) [59]. Azide- and alkyne-reporters of trehalose have also been developed for two-step labeling of mycobacteria (Fig. 8f) [60]. Moreover, a TMM chemical reporter has been developed for detection and identification of protein O-mycoloylation in Corynebacterium (Fig. 8g) [61]. In addition to hexose monosaccharide reporters, several azido-pentose reporters have been developed to label specific glycans in the mycobacterial cell wall (Fig. 8h) [62]. Moreover, arabinofuranosyl phospholipid derivatives have been generated by the Kiessling laboratory to label nucleotide-sugar labeling-resistant cell wall structures in mycobacteria [63]. Beyond individual chemical reporter labeling, multi-reporter labeling studies can help illuminate spatial and temporal features of bacterial cell division. For example, the use of orthogonal D-Ala and trehalose reporters by the Siegrist and Swarts laboratories enabled the discrimination of monopeptide-marked L,D-transpeptidase remodeling and dipeptide-marked synthesis in mycobacteria [64]. These studies highlight the unique and structurally diverse bacterial glycan biosynthetic pathways and glycoconjugates that can be labeled by chemical reporters.
Figure 8.
Chemical reporters for metabolic labeling of the mycobacterial cell envelope. (a) Schematic illustration of mycobacterial cell envelope. (b) Structures of trehalose, TMM and TDM and their interconversion by Ag85. (c) 6-FITC-Tre [58]. (d) DMN-Tre [59]. (e) QTF [8]. (f) 2-az-Tre and 6-az-Tre [60]. (g) O-Alk-TMM [61]. (h) Azido-ribose and arabinose reporters [62].
Glycans can also be directly attached to proteins in bacteria. Although protein glycosylation in bacteria shares similar biosynthetic machinery to eukaryotes, the bacteria glycans have unique and diverse structures (Fig. 9a) [65, 66]. To label these bacterial glycoproteins, the Dube laboratory synthesized several azido-monosaccharides and explored their metabolic incorporation in diverse bacterial species (Fig. 9b–d) [67]. For example, GlcNAz can only label proteins in Helicobacter pylori and Ralstonia solanacerum (Fig. 9b) [67], while azide analogs of bacillosamine (Bac-diNAz) and 2,4-diacetamido-2,4,6-trideoxygalactose (DATDG-diNAz) label proteins in H. pylori, C. jejuni, B. thailandensis and R. solanacerum (Fig. 9c) [67]. In contrast, FucNAz was not incorporated into any of these bacteria (Fig. 9d) [67]. Chemical reporters of other bacterial glycans such as pseudaminic acid by the Tanner [68] and Li [69] laboratories (Fig. 9e) as well as bacteria-specific UDP-monosaccharides by the Imperiali laboratory [70] have also been developed for labeling proteins in different bacterial species. These specific bacteria-specific monosaccharide reporters and others should provide new tools to selectively label distinct species, characterize biosynthetic enzymes and discover bacterial glycoproteins.
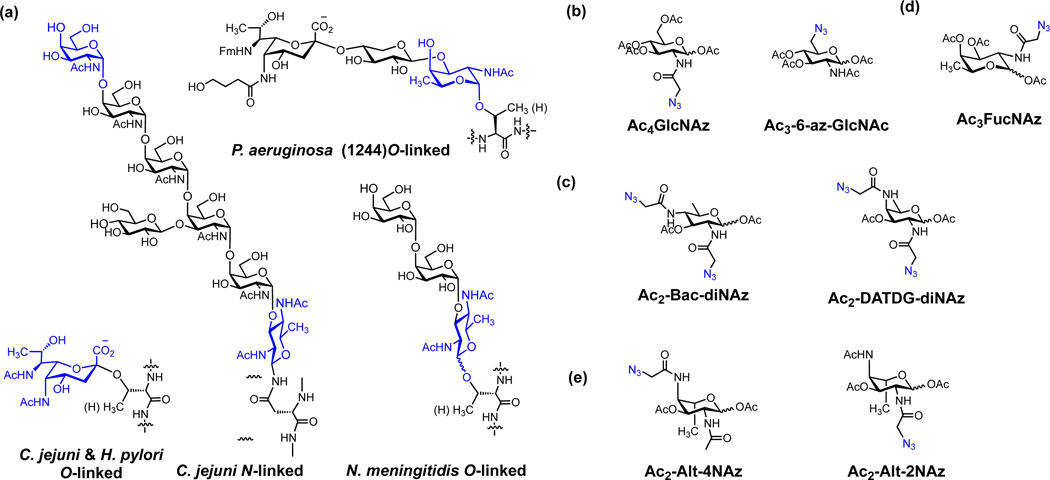
Figure 9.
Chemical reporters for labeling bacterial glycoproteins. (a) Structures of representative bacterial protein glycosylations. (b) Ac4GlcNAz and Ac3-6-az-GlcNAc [67]. (c) Ac2-Bac-diNAz and Ac2-DATDG-diNAz [67]. (d) Ac3FucNAz [67]. (e) Pseudaminic acid reporters Ac2-Alt-4NAz and Ac2-Alt-2NAz [68, 69].
Lipid reporters
Chemical reporters of lipids can also be used to label bacteria. For the imaging of membranes in bacteria, styryl dyes such as FM 4–64 are often used to stain hydrophobic lipids [71], but no specific lipid reporters have been developed for general imaging of bacteria lipids. However, our laboratory has shown that alkyne reporters of long-chain fatty acids (Fig. 10a) [10] can be metabolically incorporated into diverse bacterial species for profiling lipoproteins with canonical lipobox-motifs (Fig. 10b) as well as discovery of unpredicted-fatty-acylated proteins [72]. Moreover, alkyne-fatty acid labeling of Gram-positive bacteria by the Tate laboratory can reveal unique biosynthetic pathways such as the existence of two active signal peptidases in Clostridium difficile for lipoprotein biogenesis [73]. In addition to labeling lipids and proteins within bacteria, the application of these fatty acid reporters can reveal host regulation of secreted bacterial virulence factors as well as host substrates of secreted bacterial enzymes during infection [74–77]. Beyond covalent fatty-acylation of bacterial proteins, bi-functional fatty acid photoaffinity reporters can be used to crosslink and characterize non-covalent fatty acid interactions with proteins (Fig. 10c) [78, 79]. For example, we have employed a palmitic acid photoaffinity reporter to characterize how long-chain fatty acids attenuates Salmonella virulence (unpublished data). Our proteomic analysis revealed that long-chain fatty acids interact with many proteins in Salmonella and can directly bind HilD (unpublished data), a key transcriptional regulator of Salmonella virulence gene expression that was previously shown to be inhibited by fatty acids in vitro [80]. These studies showcase the utility of lipid reporters to discover and characterize unpredicted lipid-protein interactions in bacteria as well as during infection of host cells.
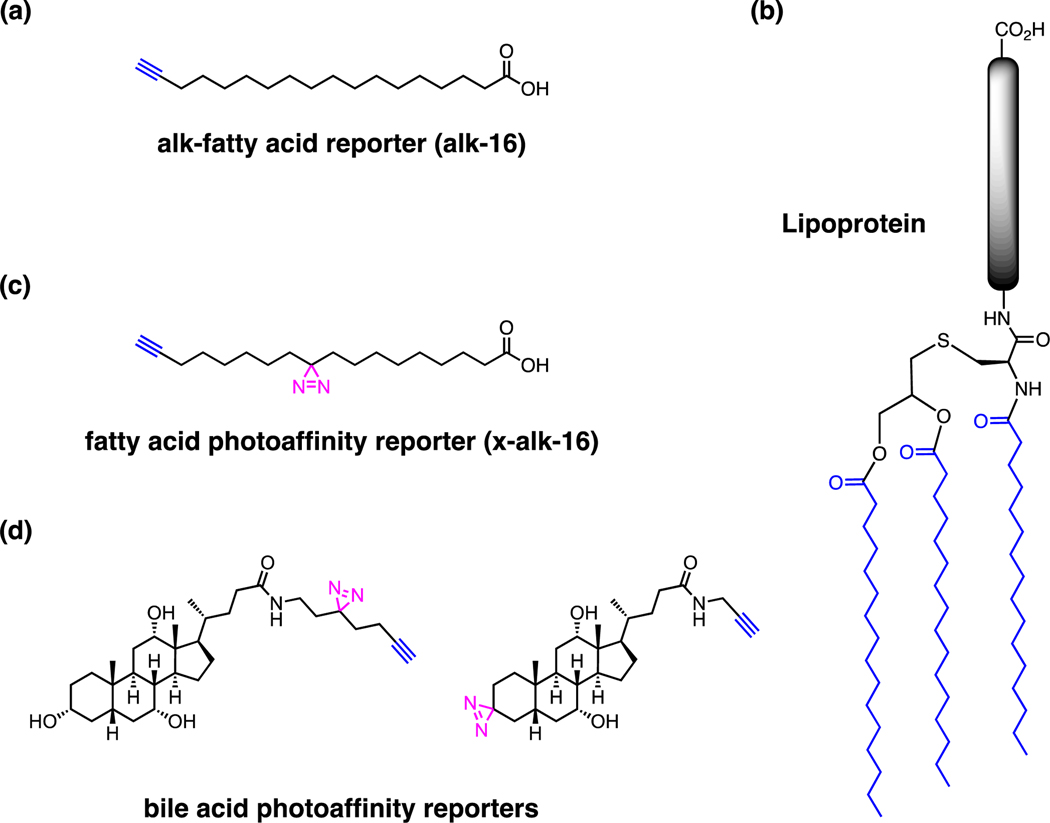
Figure 10.
Lipid-based reporters. (a) Alkyne long-chain fatty acid reporter [10, 72]. (b) Representative bacterial lipoprotein structure. (c) Fatty acid photoaffinity reporter [78]. (d) Bile acid photoaffinity reporters [81].
Activity-based reporters for microbial enzymes and metabolite targets
In addition to chemical reporters that monitor metabolite incorporation or direct binding to proteins, activity-based protein profiling (ABPP) probes or reporters have also been developed to evaluate the diverse enzymes and protein families [82, 83]. In particular, penicilin-binding proteins (PBPs) are key enyzmes that catalyze transglycosylation and transpeptidation of lipid-II subunits for peptidoglycan synthesis. In transpeptidation reaction, PBPs use their catalytic serine to form an acyl-enzyme intermediate with the peptide stem, resulting in cleavage of a D-Ala residue. A neighboring peptide stem intercepts this activated intermediate to yield crosslinked peptidoglycan. The β-lactam antibiotics covalently modify and inhibit the catalytic serine of PBPs, which the Sieber laboratory utilized to generate β-lactam reporters of PBPs (Fig. 11a) for ABPP [84, 85]. Alternatively, the Carlson laboratory has developed fluorescent and biotin derivatives of cephalosporin C (Fig. 11b), which enables selective in vivo labeling of active PBPs [86]. The Carlson laboratory has also developed β-lactone based PBP-selective reporters to distinguish the localization of PBP2x from PBP2b in S. pneumoniae (Fig. 11c) [87]. To explore other peptidoglycan biosynthetic enzymes, the Sieber laboratory has generated a chemical reporter of showdomycin (Fig. 11d) [88], a maleimide-containing uridine-based covalent inhibitor of MurA, a UDP-N-acetylglucosamine 1-carboxyvinyltransferase that catalyzes the first step in cell wall biosynthesis and antibiotic drug target. In addition, chemical reporters of acylhomoserine lactones involved in bacterial quorum sensing have been developed by the Janda laboratory to identify their potential protein targets [89]. Interestingly, the analysis of these acylhomoserine lactone reporters in mammalian cells suggests that acylhomoserine lactones may also interact with host proteins and modulate cellular pathways beyond bacteria-bacteria communication [90].
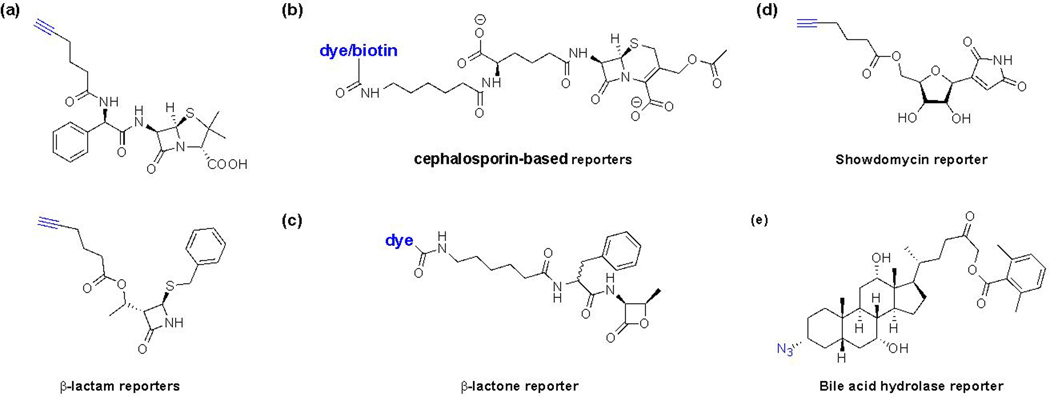
Figure 11.
Activity-based probes/reporters for microbial enzymes. (a) β-lactam-based reporters for PBPs [84, 85]. (b) Cephalosporin-based reporters for PBPs [86]. (c) β-lactone-based reporters for PBPs [87]. (d) Showdomycin-based reporter for MurA1/2 [88]. (e) BSH activity-based reporter, Ch-AOMK [91].
ABPP also provides new opportunities for discovering new antimicrobial agents. For example, the Bogyo laboratory has employed activity-based probes/reporters in combination with candidate inhibitor screening and discovered novel inhibitors of bacterial proteases that are important for the virulence of challenging and drug-resistant pathogens such as Clostridium difficile [92] and Staphylococcus aureus [93]. These select examples highlight how ABPP reporters are excellent tools for profiling of specific protein families involved fundamental microbial physiology as well as the discovery and development of new antimicrobial agents.
EXPLORATION OF MICROBIOTA MECHANISMS WITH CHEMICAL REPORTERS
The microbiota of plants, insects, animals, and humans have emerged as important factors in host physiology, disease and response to therapeutics (Fig. 1) [94–97]. These diverse communities of microbes encode unique enzymes for metabolism and also generate metabolites that can directly modulate their interactions with the host and other microbes. While microbiota composition is correlated with diverse phenotypes in metabolism, immunity and neurological function, the mechanisms of specific microbes and their metabolites have been challenging to elucidate and are only beginning to emerge [96, 98, 99]. Since the microbiota composition is influenced by host genetics and environmental factors such as diet, exposure to drugs and infections, new tools that can help determine their specific microbiota composition, spatial distribution, activities, and mechanisms with host cells and other microbes are needed. Chemical reporters of microbiota metabolites and their associated enzymes are providing new tools to address these challenges in microbiota research and are highlighted below.
Analysis of microbiota composition and dynamics
The advances in nucleic acid sequencing methods have provided important insight into microbiota composition and metagenomics in different animal models and disease states in humans. Understanding the dynamics of specific microbiota species, their spatial distribution and unique biochemical activities in vivo are key for further mechanistic studies. In this regard, the Kasper laboratory has employed monosaccharide chemical reporters to label and visualize different commensal bacteria species in vivo [44, 100]. For example, the Kasper laboratory has demonstrated that metabolic labeling of B. fragilis, B. ovatus, and B. vulgatus with GalNAz, bioorthogonal tagging with different fluorophores and recolonization of germ-free mice enabled the visualization of their unique spatial distribution in the gut in vivo [44]. They also demonstrated differential labeling of specific commensal bacteria with other monosaccharide reporters such as GlcNAz, ManNAz, and GalNAz [44]. Moreover, they showed that multi-reporter labeling of B. vulgatus with GalCCP, azido-Kdo and HADA and orthogonal fluorophores can simultaneously image the dynamics and distribution of bacteria CPS, LPS, and peptidoglycan in macrophages and the intestinal lumen of mice ex vivo [100]. In addition, the Chen laboratory has shown that azido-Kdo can be used for selective imaging, sorting, as well as 16S rRNA sequencing of Gram-negative versus Gram-positive bacteria in microbiota samples [101]. The metabolic labeling of microbiota samples with chemical reporters also provide an important means to monitor metabolic activity and viability of specific bacterial species. Indeed, chemical reporters of protein and peptidoglycan synthesis have been employed to visualize specific microbiota from environmental samples [102].
Investigation of microbiota-metabolite mechanisms
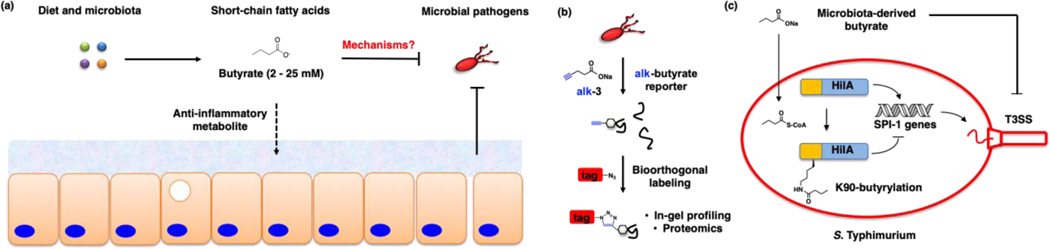
Metabolomic studies have now shown that specific microbiota species contribute to the fermentation and production of key metabolites in significant levels in the gut and circulation. For example, short-chain fatty acids (SCFAs) such as butyrate from specific microbiota (Clostridia clusters IV, XIVa, XVIII) have been shown to function as nutrients for colonocytes and suppress inflammatory pathways through GPCR activation [103, 104], inhibition of histone deacetylases (HDACs) [105] and reduction of oxidative stress [106] (Fig. 12a). Furthermore, these abundantly produced microbiota metabolites can also inhibit the expression of virulence genes in S. Typhimurium and attenuate infection, but their molecular target(s) in enteric pathogens were unknown (Fig. 12a). To identify the protein targets of SCFAs in enteric pathogens, we have employed chemical reporters of SCFA [107] (Fig. 12b) and discovered that several virulence-associated proteins encoded by the Salmonella pathogenicity island-1 (SPI-1) locus may be directly acylated (Zhang Z et al, Nat. Chem. Biol., in press). Subsequent mass spectrometry analysis, CRISPR-Cas9 gene editing and genetic code expansion revealed that site-specific modification of HilA, a transcriptional regulator of SPI-1 gene expression, attenuates S. Typhimurium invasion of epithelial cells and dissemination in vivo. Through site-specific incorporation of a stable butyryl-lysine analog that mimics gain of modification, our results showed that chemical acylation of a key residue (K90) in the DNA-binding domain of HilA may enable butyrate-producing microbiota to inhibit pathogen virulence (Fig. 12c). These results demonstrate how classical site-directed mutagenesis to canonical amino acids may not reveal how site-specific metabolite-modifications regulate protein function in cells. Moreover, our data suggests that S. Typhimurium may not only utilize butyrate as a nutrient source from the microbiota [108] but also sense high concentrations of SCFAs in the gut through HilA-acylation and other SPI-1 effectors to limit bacterial invasion of host cells. Interestingly, our analysis of medicinal plant extracts also revealed that anti-infective metabolites can covalently label and inactivate SPI-1 effector proteins in S. Typhimurium [109]. These studies showcase how chemical proteomic methods can be used to characterize unpredicted metabolite targets in pathogens, which provides new opportunities to identify and functionally characterize the molecular targets of other dietary and microbiota metabolites in the future.
Figure 12.
Microbiota-derived butyrate inhibits enteric pathogen virulence through site-specific protein acylation. (a) Microbiota-derived butyrate is a nutrient source and suppresses host inflammatory pathways as well as inhibits infection by enteric pathogens. (b) Chemical proteomic analysis of butyrate-modified proteins in S. Typhimurium. (c) Site-specific K90-butyrylation of HilA inhibits the expression of S. Typhimurium virulence genes (SPI-1), infection of epithelial cells and mice in vivo (Zhang Z et al, Nat. Chem. Biol., in press).
Beyond SCFAs, specific microbiota species have now been implicated in the production of other more complex bioactive metabolites [110]. For example, microbiota can modify long-chain fatty acids (LCFA) derived from diet, or synthesize them de novo [111]. In particular, the abundance of Lactobacillus in the gut microbiota is associated with different unsaturated and branched LCFAs in the gut and the serum of the host [112, 113]. Oral administration of a strain of Lactobacillus reuteri increases polyunsaturated LCFA serum concentration in piglets [112]. Fatty acid biosynthesis (Fab) genes are largely identified from the genera Prevotella, Lactobacillus, and Alistipes, and the excess of intracolonic saturated LCFA contributes to enhanced bowel motility in rats [113]. LCFA also directly regulates virulence of gut pathogens. For example, LCFAs have been reported to non-covalently interact with transcription regulator ToxT in Vibro cholerae [114] and HilD in Salmonella Typhimurium [80], impairing their DNA-binding ability in vitro. In addition, LCFA may act as signaling molecules through PhoP/Q two-component system to regulate SPI-1 activity in Salmonella [115]. Alternatively, other species of microbiota such as Clostridium scindens has been shown to generate secondary bile acids that may directly affect the colonization of pathogens such as C. difficile [116] or vancomycin-resistant Enterococcus faecium [117]. Moreover, the production of aromatic acids by Clostridium sporogenes has been suggested to modulate host immunity [118]. Furthermore, targeted screening of specific microbiota species and their metabolites with different protein families such as GPCRs have revealed new commensal bacteria and metabolite interactions [119–121]. These exciting studies suggest specific microbiota species may generate unique metabolites to alter host immunity or infection, but the protein targets and mechanism(s) of action for these microbiota-derived metabolites in host cells and/or microbes have not been fully characterized, thus may benefit from future studies with chemical reporters. Indeed, photoaffinity reporters have been previously generated for fatty acids (Fig. 10c) [78], cholesterol [122] and bile acids [81] (Fig. 10d) and could be readily developed for other microbiota-associated metabolites.
In addition to characterizing the direct targets of microbiota metabolites, elucidating their transporters, processing enzymes and protein targets is crucial. In this regard, ABPP using functionalized metabolites is a powerful approach for the discovery of microbial metabolite processing enzymes and their associated microbiota [83]. For example, the Wright laboratory has generated a vitamin B12-based chemical reporter that enables discovery of a light-sensing B12-binding transcription regulator that controls folate and ubiquinone biosynthesis [123]. The Wright laboratory has also generated fluorophore-modified mechanism-based probe of β-glucuronidases to label and identify specific microbiota species that contain this activity [124]. Alternatively, the Chang laboratory has generated an azide-functionalized bile salt hydrolase (BSH) reporter (Fig. 11e) to identify these enzymes from specific cultured microbiota species as well as from fecal and tissue samples [91]. For newly discovered microbiota-metabolites such as pyrazinones and dihydropyrazinones by the Fischbach laboratory, the application of activity-based probes helped determine cathepsin cysteine proteases as key host targets for these cryptic peptidic aldehydes from the microbiota [125]. These examples showcase how activity-based probes/reporters can capture microbiota-metabolite protein interactions that may not be readily apparent by the analysis of metagenomic or transcriptome datasets and will be important tools for future microbiota studies.
CONCLUDING REMARKS
Understand the chemistry of microbes is crucial for ecology and human health. This review highlights in the innovations in chemical biology, with a focus on how small molecule/metabolite reporters and bioorthogonal chemistry has helped characterize key processes in microbiology and can now be applied to more complex microbiota studies. The examples summarized above showcase the utilty of existing metabolite reporters. Nontheless, the labeling other classes of biomolecules (e.j. nucleic acids and others) in bacteria still remaining challenge and should benefit from new innovations in metabolite reporters. Furthermore, the ABPPs for other classes of proteins and synthetic fragment-based reporters could help characterize unannotated proteins bacteria and reveal new targets for antimicrobial development.
Beyond individual bacterial species, the complex microbial communities in host microbiota are now appreciated as a ‘second endocrine system’, as it produces a great variety of secondary metabolites that impact host physiology and diseases. As metagenomics and metabolomics are beginning to reveal specific composition microbiota and metabolites that are associated with health and disease, the application of chemical biology methods described above should further facilitate the mechanistic dissection of microbe-microbe and microbe-host interactions that are major challenges in microbiota studies. Given the significant associations of the microbiota with major physiological pathways in animals and humans, understanding the molecular mechanisms of microbes may provide important new targets and therapeutic leads for a variety of diseases.
ACKNOWLEDGMENT
Z.J.Z. received support from the David Rockefeller Graduate Program. Y.C.W. was a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. X.Y. received support from the Lerner Trust. H.C.H. acknowledges support from the National Institutes of Health (NIGMS R01GM103593 and R01GM087544).
Biographical Sketch

Zhenrun “Jerry” Zhang received his B.S. in Chemistry from Peking University in 2012, completing his thesis in Prof. Peng R. Chen lab. He then went on to obtain his Ph.D. at The Rockefeller University in 2018 under the guidance of Prof. Howard C. Hang. His graduate research focused on elucidating molecular mechanisms by which microbiota- and diet-derived small molecules inhibit the virulence of enteric pathogens. His work found that short-chain fatty acids chemically acylate key virulence factors in Salmonella and inhibit bacterial infection. He is currently a postdoctoral scholar at The University of Chicago.

Xinglin Yang received a B.S. in Pharmaceutical Sciences from Shandong University, China in 2012. He then obtained his PhD at Tsinghua University, working on development of novel organic synthetic methodologies towards constructing compound libraries with novelty and diversity for Drug Discovery. Currently he is a postdoctoral scholar in Hang lab at the Rockefeller University. His work involves the development of microbiota metabolite reporters to study molecular mechanism of the impact by metabolites in microbial pathogenesis.

Yen-Chih Wang received a B.S. in chemistry from National Taiwan University, Taiwan in 2006. He then completed a Ph.D. in chemistry with Prof. Mark Distefano at University of Minnesota in 2014. He then moved to the Rockefeller University as a Postdoctoral Associate under the guidance of Prof. Howard Hang. Dr. Wang’s research in the Hang lab focused on synthesizing peptidoglycan probes for enhancing host immunity against bacterial pathogens and identification of binding proteins from host cells. His research was also sponsored by CRI Irvington Postdoctoral Fellowship from 2016 to 2018.

Howard C. Hang is an Associate Professor and Head of the Laboratory of Chemical Biology and Microbial Pathogenesis at The Rockefeller University. He obtained his B.S. degree in chemistry from the University of California, Santa Cruz, 1998 with Professor Joseph P. Konopelski. In 2003, he completed his Ph.D. in chemistry at University of California, Berkeley, with Professor Carolyn Bertozzi. During his graduate studies, he was awarded an American Chemical Society, Organic Division, Graduate Fellowship. He then worked with Professor Hidde Ploegh at Harvard Medical School and the Whitehead Institute of Biomedical Research at Massachusetts Institute of Technology from 2004 through 2006 as Damon Runyon Cancer Research Foundation Postdoctoral Fellow. He joined the faculty at The Rockefeller University in 2007. He received Irma T. Hirschl Early Career Scientist Award in 2007, Ellison Foundation New Scholar in Aging Award in 2008, Distinguished Teaching Award from The Rockefeller University in 2011 and Eli Lilly Award in Biological Chemistry from American Chemical Society Division of Biological Chemistry in 2017. The Hang laboratory is broadly interested in developing chemical tools for elucidating fundamental mechanisms of host-microbe interactions and developing new therapeutic strategies towards infection, inflammation and immunotherapy.
REFERENCES
- [1].Saxon E, Bertozzi CR, Science (80-. ), 2000, 287, 2007–2010. [DOI] [PubMed] [Google Scholar]
- [2].Rostovtsev VV, Green LG, Fokin VV, Sharpless KB, Angew. Chemie - Int. Ed, 2002, 41, 2596–2599. [DOI] [PubMed] [Google Scholar]
- [3].Tornøe CW, Christensen C, Meldal M, J. Org. Chem, 2002, 67, 3057–3064. [DOI] [PubMed] [Google Scholar]
- [4].Blackman ML, Royzen M, Fox JM, J. Am. Chem. Soc, 2008, 130, 13518–13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Grammel M, Hang HC, Chemical reporters for biological discovery. Nat. Chem. Biol 9 (2013), pp. 475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li J, Chen PR, Nat. Chem. Biol, 2016, 12, 129–137. [DOI] [PubMed] [Google Scholar]
- [7].Prescher JA, Dube DH, Bertozzi CR, Nature, 2004, 430, 873–877. [DOI] [PubMed] [Google Scholar]
- [8].Hodges HL, Brown RA, Crooks JA, Weibel DB, Kiessling LL, Proc. Natl. Acad. Sci. U. S. A, 2018, 115, 5271–5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kiick KL, Saxon E, Tirrell DA, Bertozzi CR, Proc. Natl. Acad. Sci, 2002, 99, 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC, J. Am. Chem. Soc, 2009, 131, 4967–4975. [DOI] [PubMed] [Google Scholar]
- [11].Palaniappan KK, Bertozzi CR, Chem. Rev, 2016, 116, 14277–14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Roberts AM, Ward CC, Nomura DK, Curr. Opin. Biotechnol, 2017, 43, 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yount JS, Moltedo B, Yang Y-Y, Charron G, Moran TM, López CB, Hang HC, Nat. Chem. Biol, 2010, 6, 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Van Hest JCM, Kiick KL, Tirrell DA, J. Am. Chem. Soc, 2000, 122, 1282–1288. [Google Scholar]
- [15].Link AJ, Vink MKS, Tirrell DA, J. Am. Chem. Soc, 2004, doi: 10.1021/ja047629c. [DOI] [PubMed] [Google Scholar]
- [16].Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, Beatty KE, Connor RE, Yoo TH, Dieterich DC, Schuman EM, Tirrell DA, Nat. Chem. Biol, 2009, 5, 715–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Grammel M, Zhang MM, Hang HC, Angew. Chemie Int. Ed, 2010, 49, 5970–5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Grammel M, Dossa PD, Taylor-Salmon E, Hang HC, Chem. Commun, 2011, 48, 1473–4. [DOI] [PubMed] [Google Scholar]
- [19].Babin BM, Atangcho L, van Eldijk MB, Sweredoski MJ, Moradian A, Hess S, Tolker-Nielsen T, Newman DK, Tirrell DA, MBio, 2017, 8, e01593–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Silhavy TJ, Kahne D, Walker S, Cold Spring Harb. Perspect. Biol, 2010, 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Radkov AD, Hsu Y-P, Booher G, VanNieuwenhze MS, Annu. Rev. Biochem, 2018, 87, 991–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sadamoto R, Niikura K, Sears PS, Liu H, Wong CH, Suksomcheep A, Tomita F, Monde K, Nishimura SI, J. Am. Chem. Soc, 2002, 124, 9018–9019. [DOI] [PubMed] [Google Scholar]
- [23].Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, De Pedro MA, Brun YV, Vannieuwenhze MS, Angew. Chemie - Int. Ed, 2012, 51, 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, Brun YV, VanNieuwenhze MS, Vollmer W, Horn M, Jensen GJ, Nat. Commun, 2013, 4, 2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, Bertozzi CR, ACS Chem. Biol, 2013, 8, 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Shieh P, Siegrist MS, Cullen AJ, Bertozzi CR, Proc. Natl. Acad. Sci. U. S. A, 2014, 111, 5456–5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Geiger T, Pazos M, Lara-Tejero M, Vollmer W, Galán JE, Nat. Microbiol, 2018, 3, 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, Vannieuwenhze M, Maurelli AT, Nature, 2014, 506, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pidgeon SE, Pires MM, Angew. Chemie - Int. Ed, 2017, 56, 8839–8843. [DOI] [PubMed] [Google Scholar]
- [30].Pidgeon SE, Apostolos AJ, Nelson JM, Shaku M, Rimal B, Islam MN, Crick DC, Kim SJ, Pavelka MS, Kana BD, Pires MM, ACS Chem. Biol, 2019, 14, 2185–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sarkar S, Libby EA, Pidgeon SE, Dworkin J, Pires MM, Angew. Chemie Int. Ed, 2016, 55, 8401–8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liang H, DeMeester KE, Hou CW, Parent MA, Caplan JL, Grimes CL, Nat. Commun, 2017, 8, doi: 10.1038/ncomms15015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Gisin J, Schneider A, Nägele B, Borisova M, Mayer C, Nat. Chem. Biol, 2013, 9, 491–493. [DOI] [PubMed] [Google Scholar]
- [34].Borisova M, Gisin J, Mayer C, MBio, 2017, 8, doi: 10.1128/mBio.00092-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Fumeaux C, Bernhardt TG, MBio, 2017, 8, doi: 10.1128/mBio.00092-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Demeester KE, Liang H, Jensen MR, Jones ZS, D’Ambrosio EA, Scinto SL, Zhou J, Grimes CL, J. Am. Chem. Soc, 2018, 140, 9458–9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang Y, Lazor KM, DeMeester KE, Liang H, Heiss TK, Grimes CL, J. Am. Chem. Soc, 2017, 139, 13596–13599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang Y-C, Westcott NP, Griffin ME, Hang HC, ACS Chem. Biol, 2019, 14, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Whitfield C, Trent MS, Annu. Rev. Biochem, 2014, 83, 99–128. [DOI] [PubMed] [Google Scholar]
- [40].Dumont A, Malleron A, Awwad M, Dukan S, Vauzeilles B, Angew. Chemie - Int. Ed, 2012, 51, 3143–3146. [DOI] [PubMed] [Google Scholar]
- [41].Nilsson I, Grove K, Dovala D, Uehara T, Lapointe G, Six DA, J. Biol. Chem, 2017, 292, 19840–19848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yi W, Liu X, Li Y, Li J, Xia C, Zhou G, Zhang W, Zhao W, Chen X, Wang PG, Proc. Natl. Acad. Sci, 2009, 106, 4207–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pons JM, Dumont A, Sautejeau G, Fugier E, Baron A, Dukan S, Vauzeilles B, Angew. Chemie - Int. Ed, 2014, 53, 1275–1278. [DOI] [PubMed] [Google Scholar]
- [44].Geva-Zatorsky N, Alvarez D, Hudak JE, Reading NC, Erturk-Hasdemir D, Dasgupta S, Von Andrian UH, Kasper DL, Nat. Med, 2015, 21, 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stenutz R, Weintraub A, Widmalm G, The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev 30 (2006), pp. 382–403. [DOI] [PubMed] [Google Scholar]
- [46].Liu B, Knirel YA, Feng L, Perepelov AV, Senchenkova SN, Reeves PR, Wang L, Structural diversity in Salmonella O antigens and its genetic basis. FEMS Microbiol. Rev 38 (2014), pp. 56–89. [DOI] [PubMed] [Google Scholar]
- [47].Ratcliff RM, Lanser JA, Manning PA, Heuzenroeder MW, J. Clin. Microbiol, 1998, 36, 1560–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Knirel YA, Senchenkova SN, Kocharova NA, Shashkov AS, Helbig JH, Zähringer U, Biochem, 2001, 66, 1035–1041. [DOI] [PubMed] [Google Scholar]
- [49].Whitfield C, Annu. Rev. Biochem, 2006, 75, 39–68. [DOI] [PubMed] [Google Scholar]
- [50].Yother J, Annu. Rev. Microbiol, 2010, 65, 563–581. [DOI] [PubMed] [Google Scholar]
- [51].Mazmanian SK, Round JL, Kasper DL, Nature, 2008, 453, 620–5. [DOI] [PubMed] [Google Scholar]
- [52].Orskov I, Orskov F, Jann B, Jann K, Bacteriol. Rev, 1977, 41, 667–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].van Dam JEG, Fleer A, Snippe H, Immunogenicity and immunochemistry of Streptococcus pneumoniae capsular polysaccharides. Antonie Van Leeuwenhoek. 58 (1990), pp. 1–47. [DOI] [PubMed] [Google Scholar]
- [54].Jankute M, Cox JAG, Harrison J, Besra GS, Annu. Rev. Microbiol, 2015, 69, 405–423. [DOI] [PubMed] [Google Scholar]
- [55].Backus KM, Boshoff HI, Barry CS, Boutureira O, Patel MK, D’Hooge F, Lee SS, Via LE, Tahlan K, Barry CE, Davis BG, Nat. Chem. Biol, 2011, 7, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Swarts BM, Holsclaw CM, Jewett JC, Alber M, Fox DM, Siegrist MS, Leary JA, Kalscheuer R, Bertozzi CR, J. Am. Chem. Soc, 2012, 134, 16123–16126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Foley HN, Stewart JA, Kavunja HW, Rundell SR, Swarts BM, Angew. Chemie - Int. Ed, 2016, 55, 2053–2057. [DOI] [PubMed] [Google Scholar]
- [58].Rodriguez-Rivera FP, Zhou X, Theriot JA, Bertozzi CR, J. Am. Chem. Soc, 2017, 139, 3488–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kamariza M, Shieh P, Ealand CS, Peters JS, Chu B, Rodriguez-Rivera FP, Babu Sait MR, V Treuren W, Martinson N, Kalscheuer R, Kana BD, Bertozzi CR, Sci. Transl. Med, 2018, 10, eaam6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Fiolek TJ, Banahene N, Kavunja HW, Holmes NJ, Rylski AK, Pohane AA, Siegrist MS, Swarts BM, ChemBioChem, 2019, 20, 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kavunja HW, Piligian BF, Fiolek TJ, Foley HN, Nathan TO, Swarts BM, Chem. Commun, 2016, 52, 13795–13798. [DOI] [PubMed] [Google Scholar]
- [62].Kolbe K, Möckl L, Sohst V, Brandenburg J, Engel R, Malm S, Bräuchle C, Holst O, Lindhorst TK, Reiling N, ChemBioChem, 2017, 18, 1172–1176. [DOI] [PubMed] [Google Scholar]
- [63].Calabretta PJ, Hodges HL, Kraft MB, Marando VM, Kiessling LL, J. Am. Chem. Soc, 2019, 141, 9262–9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].García-Heredia A, Pohane AA, Melzer ES, Carr CR, Fiolek TJ, Rundell SR, Lim HC, Wagner JC, Morita YS, Swarts BM, Siegrist MS, Elife, 2018, 7, doi: 10.7554/eLife.37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Dube DH, Champasa K, Wang B, Chemical tools to discover and target bacterial glycoproteins. Chem. Commun 47 (2011), pp. 87–101. [DOI] [PubMed] [Google Scholar]
- [66].Tra VN, Dube DH, Chem. Commun, 2014, 50, 4659–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Clark EL, Emmadi M, Krupp KL, Podilapu AR, Helble JD, Kulkarni SS, Dube DH, ACS Chem. Biol, 2016, 11, 3365–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liu F, Aubry AJ, Schoenhofen IC, Logan SM, Tanner ME, ChemBioChem, 2009, 10, 1317–1320. [DOI] [PubMed] [Google Scholar]
- [69].Andolina G, Wei R, Liu H, Zhang Q, Yang X, Cao H, Chen S, Yan A, Li XD, Li X, ACS Chem. Biol, 2018, 13, 3030–3037. [DOI] [PubMed] [Google Scholar]
- [70].Lukose V, Whitworth G, Guan Z, Imperiali B, J. Am. Chem. Soc, 2015, 137, 12446–12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Fishov I, Woldringh CL, Mol. Microbiol, 1999, 32, 1166–1172. [DOI] [PubMed] [Google Scholar]
- [72].Rangan KJ, Yang Y-Y, Charron G, Hang HC, J. Am. Chem. Soc, 2010, 132, 10628–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Charlton TM, Kovacs-Simon A, Michell SL, Fairweather NF, Tate EW, Chem. Biol, 2015, 22, 1562–1573. [DOI] [PubMed] [Google Scholar]
- [74].Burnaevskiy N, Peng T, Reddick LE, Hang HC, Alto NM, Mol. Cell, 2015, 58, 110–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Burnaevskiy N, Fox TG, Plymire DA, Ertelt JM, Weigele BA, Selyunin AS, Way SS, Patrie SM, Alto NM, Nature, 2013, 496, 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhou Y, Huang C, Yin L, Wan M, Wang X, Li L, Liu Y, Wang Z, Fu P, Zhang N, Chen S, Liu X, Shao F, Zhu Y, Science, 2017, 358, 528–531. [DOI] [PubMed] [Google Scholar]
- [77].Liu W, Zhou Y, Peng T, Zhou P, Ding X, Li Z, Zhong H, Xu Y, Chen S, Hang HC, Shao F, Nat. Microbiol, 2018, 3, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Peng T, Hang HC, J. Am. Chem. Soc, 2015, 137, 556–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Haberkant P, Raijmakers R, Wildwater M, Sachsenheimer T, Brügger B, Maeda K, Houweling M, Gavin AC, Schultz C, Van Meer G, Heck AJR, Holthuis JCM, Angew. Chemie - Int. Ed, 2013, 52, 4033–4038. [DOI] [PubMed] [Google Scholar]
- [80].Golubeva YA, Ellermeier JR, Cott Chubiz JE, Slauch JM, Chubiz JEC, Slauch JM, MBio, 2016, 7, e02170–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhuang S, Li Q, Cai L, Wang C, Lei X, ACS Cent. Sci, 2017, 3, 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Cravatt BF, Wright AT, Kozarich JW, Annu. Rev. Biochem, 2008, 77, 383–414. [DOI] [PubMed] [Google Scholar]
- [83].Sadler NC, Wright AT, Activity-based protein profiling of microbes. Curr. Opin. Chem. Biol 24 (2015), pp. 139–144. [DOI] [PubMed] [Google Scholar]
- [84].Staub I, Sieber SA, J. Am. Chem. Soc, 2008, 130, 13400–13409. [DOI] [PubMed] [Google Scholar]
- [85].Staub I, Sieber SA, J. Am. Chem. Soc, 2009, 131, 6271–6276. [DOI] [PubMed] [Google Scholar]
- [86].Kocaoglu O, Calvo RA, Sham LT, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, Carlson EE, ACS Chem. Biol, 2012, 7, 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Sharifzadeh S, Boersma MJ, Kocaoglu O, Shokri A, Brown CL, Shirley JD, Winkler ME, Carlson EE, ACS Chem. Biol, 2017, 12, 2849–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Böttcher T, Sieber SA, J. Am. Chem. Soc, 2010, 132, 6964–6972. [DOI] [PubMed] [Google Scholar]
- [89].Dubinsky L, Jarosz LM, Amara N, Krief P, Kravchenko VV, Krom BP, Meijler MM, Chem. Commun, 2009, 7378–7380. [DOI] [PubMed] [Google Scholar]
- [90].Garner AL, Yu J, Struss AK, Lowery CA, Zhu J, Kim SK, Park J, Mayorov AV, Kaufmann GF, Kravchenko VV, Janda KD, Bioorg. Med. Chem. Lett, 2011, 21, 2702–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Parasar B, Zhou H, Xiao X, Shi Q, Brito IL, Chang PV, ACS Cent. Sci, in press, doi: 10.1021/acscentsci.9b00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bender KO, Garland M, Ferreyra JA, Hryckowian AJ, Child MA, Puri AW, Solow-Cordero DE, Higginbottom SK, Segal E, Banaei N, Shen A, Sonnenburg JL, Bogyo M, Sci. Transl. Med, 2015, 7, 306ra148–306ra148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lentz CS, Sheldon JR, Crawford LA, Cooper R, Garland M, Amieva MR, Weerapana E, Skaar EP, Bogyo M, Nat. Chem. Biol, 2018, 14, 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Buffie CG, Pamer EG, Nat. Rev. Immunol, 2013, 13, 790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Lam KN, Alexander M, Turnbaugh PJ, Cell Host Microbe, 2019, 26, 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Fischbach MA, Cell, 2018, 174, 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Chen YE, Fischbach MA, Belkaid Y, Nature, 2018, 553, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Rangan KJ, Pedicord VA, Wang YC, Kim B, Lu Y, Shaham S, Mucida D, Hang HC, Science (80-. ), 2016, 353, 1434–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Pedicord VA, Lockhart AAK, Rangan KJ, Craig JW, Loschko J, Rogoz A, Hang HC, Mucida D, Sci. Immunol, 2016, 1, eaai7732–eaai7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Hudak JE, Alvarez D, Skelly A, Von Andrian UH, Kasper DL, Nat. Microbiol, 2017, 2, doi: 10.1038/nmicrobiol.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Wang W, Zhu Y, Chen X, Biochemistry, 2017, 56, 3889–3893. [DOI] [PubMed] [Google Scholar]
- [102].Hatzenpichler R, Scheller S, Tavormina PL, Babin BM, Tirrell DA, Orphan VJ, Environ. Microbiol, 2014, 16, 2568–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, Mackay CR, Nat. Commun, 2015, 6, doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- [104].Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V, Immunity, 2014, 40, 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chang PV, Hao L, Offermanns S, Medzhitov R, Proc. Natl. Acad. Sci, 2014, 111, 2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Rivera-Chávez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, Bäumler AJ, Cell Host Microbe, 2016, 19, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Yang Y-Y, Ascano JM, Hang HC, J. Am. Chem. Soc, 2010, 132, 3640–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Campbell JW, Morgan-Kiss RM, Cronan JE, Mol. Microbiol, 2003, doi: 10.1046/j.1365-2958.2003.03341.x. [DOI] [PubMed] [Google Scholar]
- [109].Tsou LK, Lara-Tejero MM, RoseFigura J, Zhang ZJ, Wang YC, Yount JS, Lefebre M, Dossa PD, Kato J, Guan F, Lam W, Cheng YC, Galán JE, Hang HC, Galan JE, Hang HC, J. Am. Chem. Soc, in press, doi: 10.1021/jacs.5b11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Donia MS, Fischbach MA, Science (80-. ), 2015, 349, 1254766–1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Grininger M, Heil CS, Wehrheim SS, Paithankar KS, ChemBioChem, in press, doi: 10.1002/cbic.201800809. [DOI] [PubMed] [Google Scholar]
- [112].Zhang D, Liu H, Wang S, Zhang W, Wang J, Tian H, Wang Y, Ji H, Front. Microbiol, 2019, 10, doi: 10.3389/fmicb.2019.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Zhao L, Huang Y, Lu L, Yang W, Huang T, Lin Z, Lin C, Kwan H, Wong HLX, Chen Y, Sun S, Xie X, Fang X, Yang H, Wang J, Zhu L, Bian Z, Microbiome, 2018, 6, doi: 10.1186/s40168-018-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ, Proc. Natl. Acad. Sci, 2010, 107, 2860–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Viarengo G, Sciara MI, Salazar MO, Kieffer PM, Furlán RLE, Véscovi EG, J. Biol. Chem, 2013, 288, 22346–22358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, Van Den Brink MRM, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG, Nature, 2015, 517, 205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].McKenney PT, Yan J, Vaubourgeix J, Becattini S, Lampen N, Motzer A, Larson PJ, Dannaoui D, Fujisawa S, Xavier JB, Pamer EG, Cell Host Microbe, 2019, 25, 695–705.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, Sonnenburg JL, Nature, 2017, 551, 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Colosimo DA, Kohn JA, Luo PM, Cross JR, Cohen LJ, Correspondence SFB, Cell Host Microbe, 2019, 26, 273–282.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Cohen LJ, Esterhazy D, Kim S-H, Lemetre C, Aguilar RR, Gordon EA, Pickard AJ, Cross JR, Emiliano AB, Han SM, Chu J, Vila-Farres X, Kaplitt J, Rogoz A, Calle PY, Hunter C, Bitok JK, Brady SF, Nature, 2017, 549, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Chen H, Nwe P-K, Yang Y, Rosen CE, Bielecka AA, Kuchroo M, Cline GW, Kruse AC, Ring AM, Crawford JM, Palm NW, Cell, 2019, 177, 1217–1231.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Budelier MM, Cheng WWL, Bergdoll L, Chen Z-W, Janetka JW, Abramson J, Krishnan K, Mydock-McGrane L, Covey DF, Whitelegge JP, Evers AS, J. Biol. Chem, 2017, 292, 9294–9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Romine MF, Rodionov DA, Maezato Y, Anderson LN, Nandhikonda P, Rodionova IA, Carre A, Li X, Xu C, Clauss TRW, Kim YM, Metz TO, Wright AT, Proc. Natl. Acad. Sci. U. S. A, 2017, 114, E1205–E1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Whidbey C, Sadler NC, Nair RN, Volk RF, DeLeon AJ, Bramer LM, Fansler SJ, Hansen JR, Shukla AK, Jansson JK, Thrall BD, Wright AT, J. Am. Chem. Soc, 2019, 141, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Guo CJ, Chang FY, Wyche TP, Backus KM, Acker TM, Funabashi M, Taketani M, Donia MS, Nayfach S, Pollard KS, Craik CS, Cravatt BF, Clardy J, Voigt CA, Fischbach MA, Cell, 2017, 168, 517–526.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]