Figure 1.
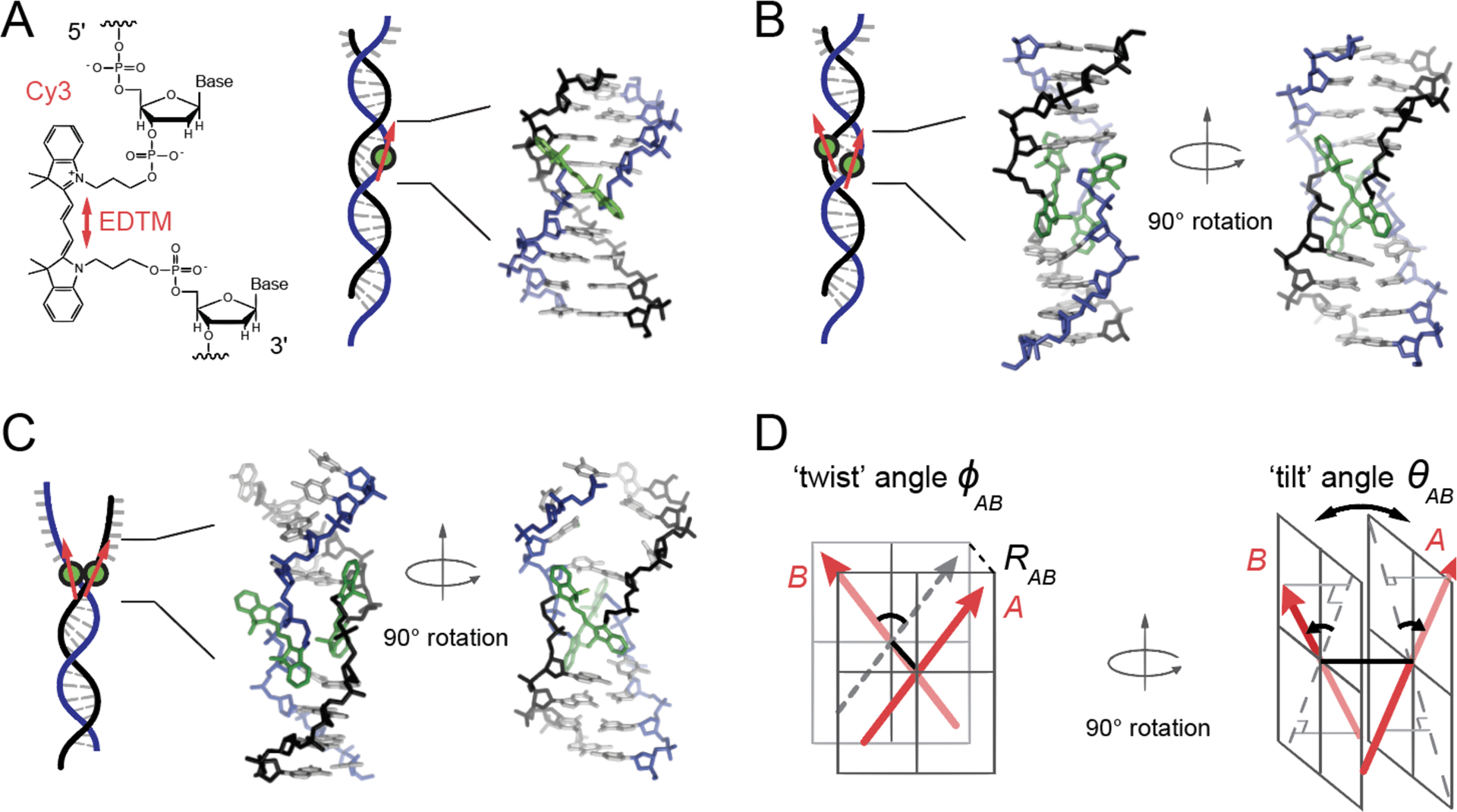
Model structure of the internally labeled Cy3 monomer and (Cy3)2 dimer in dsDNA. (A) The structural formula of the internally labeled Cy3 chromophore is shown with its insertion linkages to the 3’ and 5’ segments of the sugar-phosphate backbone of ssDNA. A red double-headed arrow indicates the orientation of the electric dipole transition moment (EDTM), which lies parallel to the axis of the trimethine bridge in the all-trans configuration. (B) A (Cy3)2 dimer-dsDNA construct is formed by annealing two complementary DNA strands, which each contain a site-specifically positioned Cy3 chromophore. Space-filling structural models performed using the Spartan program (Wavefunction, Inc.) suggest that the dimer exhibits the same approximate D2 symmetry as right-handed (B-form) helical dsDNA (17). (C) A (Cy3)2 dimer-fork DNA construct contains the dimer probe near the ss-ds DNA fork junction. The local conformation of the (Cy3)2 dimer probe is expected to reflect the disruption of base-stacking interactions that occurs near the fork junction. In panels (A – C), the sugar-phosphate backbones of the conjugate strands are shown in black and blue, the bases in gray, and the Cy3 chromophores in green. (D) The structural parameters that define the local conformation of the (Cy3)2 dimer are the inter-chromophore separation vector RAB, the twist angle ϕAB, and the tilt angle θAB.
