Abstract
The Panel on Plant Health performed a pest categorisation of non‐European isolates of Citrus tristeza virus (CTV) for the EU territory. CTV is a well characterised virus for which efficient detection assays are available. It is transmitted by vegetative multiplication of infected hosts and by aphid vectors. The most efficient one, Toxoptera citricida, has limited EU presence but another one, Aphis gossypii, is broadly distributed. CTV is reported from a range of countries outside the EU and EU isolates are present in seven of the eight citrus‐growing member states. Non‐EU isolates are not known to occur in the EU and therefore do not meet one of the criteria for being a Union regulated non‐quarantine pest. The natural host range of CTV is restricted to Citrus, Fortunella and Poncirus species. CTV non‐EU isolates are listed in Annex IIAI of Directive 2000/29/EC and the main pathway for entry, plants for planting, is closed by the existing legislation. CTV isolates may therefore only enter through minor alternative pathways. They have the potential to subsequently spread through plants for planting and through the action of aphid vectors. CTV non‐EU isolates are able to cause severe symptoms on a range of citrus crops that EU isolates do not induce. Overall, non‐EU CTV isolates meet all the criteria evaluated by EFSA to qualify as Union quarantine pests. The main knowledge gaps and uncertainties concern (1) the status of Rutaceae species other than Citrus, Fortunella and Poncirus as natural hosts for CTV; (2) the potential undetected presence of non‐EU CTV isolates in the EU and in particular the prevalence and biological properties of CTV isolates that may be present in ornamental citrus; and (3) the inability of EU CTV isolates apparently related to non‐European stem pitting (SP) isolates to cause SP in sweet orange.
Keywords: citrus tristeza virus (no‐EU), Citrus, Fortunella, Poncirus, Toxoptera citricida, Aphis gossypii, stem pitting
1. Introduction
1.1. Background and Terms of Reference as provided by the requestor
1.1.1. Background
Council Directive 2000/29/EC1 on protective measures against the introduction into the Community of organisms harmful to plants or plant products and against their spread within the Community establishes the present European Union plant health regime. The Directive lays down the phytosanitary provisions and the control checks to be carried out at the place of origin on plants and plant products destined for the Union or to be moved within the Union. In the Directive's 2000/29/EC annexes, the list of harmful organisms (pests) whose introduction into or spread within the Union is prohibited, is detailed together with specific requirements for import or internal movement.
Following the evaluation of the plant health regime, the new basic plant health law, Regulation (EU) 2016/20312 on protective measures against pests of plants, was adopted on 26 October 2016 and will apply from 14 December 2019 onwards, repealing Directive 2000/29/EC. In line with the principles of the above mentioned legislation and the follow‐up work of the secondary legislation for the listing of EU regulated pests, EFSA is requested to provide pest categorizations of the harmful organisms included in the annexes of Directive 2000/29/EC, in the cases where recent pest risk assessment/ pest categorisation is not available.
1.1.2. Terms of reference
EFSA is requested, pursuant to Article 22(5.b) and Article 29(1) of Regulation (EC) No 178/20023, to provide scientific opinion in the field of plant health.
EFSA is requested to prepare and deliver a pest categorisation (step 1 analysis) for each of the regulated pests included in the appendices of the annex to this mandate. The methodology and template of pest categorisation have already been developed in past mandates for the organisms listed in Annex II Part A Section II of Directive 2000/29/EC. The same methodology and outcome is expected for this work as well.
The list of the harmful organisms included in the annex to this mandate comprises 133 harmful organisms or groups. A pest categorisation is expected for these 133 pests or groups and the delivery of the work would be stepwise at regular intervals through the year as detailed below. First priority covers the harmful organisms included in Appendix 1, comprising pests from Annex II Part A Section I and Annex II Part B of Directive 2000/29/EC. The delivery of all pest categorisations for the pests included in Appendix 1 is June 2018. The second priority is the pests included in Appendix 2, comprising the group of Cicadellidae (non‐EU) known to be vector of Pierce's disease (caused by Xylella fastidiosa), the group of Tephritidae (non‐EU), the group of potato viruses and virus‐like organisms, the group of viruses and virus‐like organisms of Cydonia Mill., Fragaria L., Malus Mill., Prunus L., Pyrus L., Ribes L., Rubus L. and Vitis L.. and the group of Margarodes (non‐EU species). The delivery of all pest categorisations for the pests included in Appendix 2 is end 2019. The pests included in Appendix 3 cover pests of Annex I part A section I and all pests categorisations should be delivered by end 2020.
For the above mentioned groups, each covering a large number of pests, the pest categorisation will be performed for the group and not the individual harmful organisms listed under “such as” notation in the Annexes of the Directive 2000/29/EC. The criteria to be taken particularly under consideration for these cases, is the analysis of host pest combination, investigation of pathways, the damages occurring and the relevant impact.
Finally, as indicated in the text above, all references to ‘non‐European’ should be avoided and replaced by ‘non‐EU’ and refer to all territories with exception of the Union territories as defined in Article 1 point 3 of Regulation (EU) 2016/2031.
1.1.2.1. Terms of Reference: Appendix 1
List of harmful organisms for which pest categorisation is requested. The list below follows the annexes of Directive 2000/29/EC.
| Annex IIAI | |
| (a) Insects, mites and nematodes, at all stages of their development | |
| Aleurocantus spp. | Numonia pyrivorella (Matsumura) |
| Anthonomus bisignifer (Schenkling) | Oligonychus perditus Pritchard and Baker |
| Anthonomus signatus (Say) | Pissodes spp. (non‐EU) |
| Aschistonyx eppoi Inouye | Scirtothrips aurantii Faure |
| Carposina niponensis Walsingham | Scirtothrips citri (Moultex) |
| Enarmonia packardi (Zeller) | Scolytidae spp. (non‐EU) |
| Enarmonia prunivora Walsh | Scrobipalpopsis solanivora Povolny |
| Grapholita inopinata Heinrich | Tachypterellus quadrigibbus Say |
| Hishomonus phycitis | Toxoptera citricida Kirk. |
| Leucaspis japonica Ckll. | Unaspis citri Comstock |
| Listronotus bonariensis (Kuschel) | |
| (b) Bacteria | |
| Citrus variegated chlorosis | Xanthomonas campestris pv. oryzae (Ishiyama) Dye and pv. oryzicola (Fang. et al.) Dye |
| Erwinia stewartii (Smith) Dye | |
| (c) Fungi | |
| Alternaria alternata (Fr.) Keissler (non‐EU pathogenic isolates) | Elsinoe spp. Bitanc. and Jenk. Mendes |
| Anisogramma anomala (Peck) E. Müller | Fusarium oxysporum f. sp. albedinis (Kilian and Maire) Gordon |
| Apiosporina morbosa (Schwein.) v. Arx | Guignardia piricola (Nosa) Yamamoto |
| Ceratocystis virescens (Davidson) Moreau | Puccinia pittieriana Hennings |
| Cercoseptoria pini‐densiflorae (Hori and Nambu) Deighton | Stegophora ulmea (Schweinitz: Fries) Sydow & Sydow |
| Cercospora angolensis Carv. and Mendes | Venturia nashicola Tanaka and Yamamoto |
| (d) Virus and virus‐like organisms | |
| Beet curly top virus (non‐EU isolates) | Little cherry pathogen (non‐ EU isolates) |
| Black raspberry latent virus | Naturally spreading psorosis |
| Blight and blight‐like | Palm lethal yellowing mycoplasm |
| Cadang‐Cadang viroid | Satsuma dwarf virus |
| Citrus tristeza virus (non‐EU isolates) | Tatter leaf virus |
| Leprosis | Witches’ broom (MLO) |
| Annex IIB | |
| (a) Insect mites and nematodes, at all stages of their development | |
| Anthonomus grandis (Boh.) | Gilphinia hercyniae (Hartig) |
| Cephalcia lariciphila (Klug) | Gonipterus scutellatus Gyll. |
| Dendroctonus micans Kugelan | Ips amitinus Eichhof |
| Ips cembrae Heer | Ips typographus Heer |
| Ips duplicatus Sahlberg | Sternochetus mangiferae Fabricius |
| Ips sexdentatus Börner | |
| (b) Bacteria | |
| Curtobacterium flaccumfaciens pv. flaccumfaciens (Hedges) Collins and Jones | |
| (c) Fungi | |
| Glomerella gossypii Edgerton | Hypoxylon mammatum (Wahl.) J. Miller |
| Gremmeniella abietina (Lag.) Morelet | |
1.1.2.2. Terms of Reference: Appendix 2
List of harmful organisms for which pest categorisation is requested per group. The list below follows the categorisation included in the annexes of Directive 2000/29/EC.
| Annex IAI | |
| (a) Insects, mites and nematodes, at all stages of their development | |
| Group of Cicadellidae (non‐EU) known to be vector of Pierce's disease (caused by Xylella fastidiosa), such as: | |
| 1) Carneocephala fulgida Nottingham | 3) Graphocephala atropunctata (Signoret) |
| 2) Draeculacephala minerva Ball | |
| Group of Tephritidae (non‐EU) such as: | |
| 1) Anastrepha fraterculus (Wiedemann) | 12) Pardalaspis cyanescens Bezzi |
| 2) Anastrepha ludens (Loew) | 13) Pardalaspis quinaria Bezzi |
| 3) Anastrepha obliqua Macquart | 14) Pterandrus rosa (Karsch) |
| 4) Anastrepha suspensa (Loew) | 15) Rhacochlaena japonica Ito |
| 5) Dacus ciliatus Loew | 16) Rhagoletis completa Cresson |
| 6) Dacus curcurbitae Coquillet | 17) Rhagoletis fausta (Osten‐Sacken) |
| 7) Dacus dorsalis Hendel | 18) Rhagoletis indifferens Curran |
| 8) Dacus tryoni (Froggatt) | 19) Rhagoletis mendax Curran |
| 9) Dacus tsuneonis Miyake | 20) Rhagoletis pomonella Walsh |
| 10) Dacus zonatus Saund. | 21) Rhagoletis suavis (Loew) |
| 11) Epochra canadensis (Loew) | |
| (c) Viruses and virus‐like organisms | |
| Group of potato viruses and virus‐like organisms such as: | |
| 1) Andean potato latent virus | 4) Potato black ringspot virus |
| 2) Andean potato mottle virus | 5) Potato virus T |
| 3) Arracacha virus B, oca strain | 6) non‐EU isolates of potato viruses A, M, S, V, X and Y (including Yo, Yn and Yc) and Potato leafroll virus |
| Group of viruses and virus‐like organisms of Cydonia Mill., Fragaria L., Malus Mill., Prunus L., Pyrus L., Ribes L.,Rubus L. and Vitis L., such as: | |
| 1) Blueberry leaf mottle virus | 6) Peach rosette mycoplasm |
| 2) Cherry rasp leaf virus (American) | 7) Peach X‐disease mycoplasm |
| 3) Peach mosaic virus (American) | 8) Peach yellows mycoplasm |
| 4) Peach phony rickettsia | 9) Plum line pattern virus (American) |
| 5) Peach rosette mosaic virus | 10) Raspberry leaf curl virus (American) |
| 11) Strawberry witches’ broom mycoplasma | 12) Non‐EU viruses and virus‐like organisms of Cydonia Mill., Fragaria L., Malus Mill., Prunus L., Pyrus L., Ribes L., Rubus L. and Vitis L. |
| Annex IIAI | |
| (a) Insects, mites and nematodes, at all stages of their development | |
| Group of Margarodes (non‐EU species) such as: | |
| 1) Margarodes vitis (Phillipi) | 3) Margarodes prieskaensis Jakubski |
| 2) Margarodes vredendalensis de Klerk | |
1.1.2.3. Terms of Reference: Appendix 3
List of harmful organisms for which pest categorisation is requested. The list below follows the annexes of Directive 2000/29/EC.
| Annex IAI | |
| (a) Insects, mites and nematodes, at all stages of their development | |
| Acleris spp. (non‐EU) | Longidorus diadecturus Eveleigh and Allen |
| Amauromyza maculosa (Malloch) | Monochamus spp. (non‐EU) |
| Anomala orientalis Waterhouse | Myndus crudus Van Duzee |
| Arrhenodes minutus Drury | Nacobbus aberrans (Thorne) Thorne and Allen |
| Choristoneura spp. (non‐EU) | Naupactus leucoloma Boheman |
| Conotrachelus nenuphar (Herbst) | Premnotrypes spp. (non‐EU) |
| Dendrolimus sibiricus Tschetverikov | Pseudopityophthorus minutissimus (Zimmermann) |
| Diabrotica barberi Smith and Lawrence | Pseudopityophthorus pruinosus (Eichhoff) |
| Diabrotica undecimpunctata howardi Barber | Scaphoideus luteolus (Van Duzee) |
| Diabrotica undecimpunctata undecimpunctata Mannerheim | Spodoptera eridania (Cramer) |
| Diabrotica virgifera zeae Krysan & Smith | Spodoptera frugiperda (Smith) |
| Diaphorina citri Kuway | Spodoptera litura (Fabricus) |
| Heliothis zea (Boddie) | Thrips palmi Karny |
| Hirschmanniella spp., other than Hirschmanniella gracilis (de Man) Luc and Goodey | Xiphinema americanum Cobb sensu lato (non‐EU populations) |
| Liriomyza sativae Blanchard | Xiphinema californicum Lamberti and Bleve‐Zacheo |
| (b) Fungi | |
| Ceratocystis fagacearum (Bretz) Hunt | Mycosphaerella larici‐leptolepis Ito et al. |
| Chrysomyxa arctostaphyli Dietel | Mycosphaerella populorum G. E. Thompson |
| Cronartium spp. (non‐EU) | Phoma andina Turkensteen |
| Endocronartium spp. (non‐EU) | Phyllosticta solitaria Ell. and Ev. |
| Guignardia laricina (Saw.) Yamamoto and Ito | Septoria lycopersici Speg. var. malagutii Ciccarone and Boerema |
| Gymnosporangium spp. (non‐EU) | Thecaphora solani Barrus |
| Inonotus weirii (Murril) Kotlaba and Pouzar | Trechispora brinkmannii (Bresad.) Rogers |
| Melampsora farlowii (Arthur) Davis | |
| (c) Viruses and virus‐like organisms | |
| Tobacco ringspot virus | Pepper mild tigré virus |
| Tomato ringspot virus | Squash leaf curl virus |
| Bean golden mosaic virus | Euphorbia mosaic virus |
| Cowpea mild mottle virus | Florida tomato virus |
| Lettuce infectious yellows virus | |
| (d) Parasitic plants | |
| Arceuthobium spp. (non‐EU) | |
| Annex IAII | |
| (a) Insects, mites and nematodes, at all stages of their development | |
| Meloidogyne fallax Karssen | Rhizoecus hibisci Kawai and Takagi |
| Popillia japonica Newman | |
| (b) Bacteria | |
| Clavibacter michiganensis (Smith) Davis et al. ssp. sepedonicus (Spieckermann and Kotthoff) Davis et al. | Ralstonia solanacearum (Smith) Yabuuchi et al. |
| (c) Fungi | |
| Melampsora medusae Thümen | Synchytrium endobioticum (Schilbersky) Percival |
| Annex I B | |
| (a) Insects, mites and nematodes, at all stages of their development | |
| Leptinotarsa decemlineata Say | Liriomyza bryoniae (Kaltenbach) |
| (b) Viruses and virus‐like organisms | |
| Beet necrotic yellow vein virus | |
1.2. Interpretation of the Terms of Reference
Citrus tristeza virus (CTV; non‐European isolates) is one of a number of pests listed in the Appendices to the Terms of Reference (ToR) to be subject to pest categorisation to determine whether it fulfils the criteria of a quarantine pest or those of a regulated non‐quarantine pest (RNQP) for the area of the European Union (EU) excluding Ceuta, Melilla and the outermost regions of Member States referred to in Article 355(1) of the Treaty on the Functioning of the European Union (TFEU), other than Madeira and the Azores.
This pest categorisation covers non‐European isolates of CTV, which are defined by their geographical origin outside of the European Union territory. As such, CTV isolates occurring outside of the EU territory are considered as non‐EU isolates of CTV. In the same way, a plant infected with CTV originating in a non‐EU country is considered to be infected with a non‐EU CTV isolate. EU CTV isolates are not covered by the present pest categorisation, unless considered necessary for a better understanding. In this case, the extension of coverage to EU isolates is explicitly stated. However, EU isolates of CTV have been addressed in a previous Opinion of the Plant Health Panel of EFSA (EFSA, 2014).
2. Data and methodologies
2.1. Data
2.1.1. Literature search
A literature search on Citrus tristeza virus (non‐European isolates) was conducted at the beginning of the categorisation in the ISI Web of Science bibliographic database and further references and information were obtained from experts, from citations within the references and grey literature.
During its categorisation of EU CTV isolates (EFSA PLH Panel, 2014), the Plant Health panel conducted an extensive analysis of the information available on the intraspecific molecular and biological diversity of CTV. Although many new genomic sequences of CTV isolates have been reported since then, these sequences do not modify in any major way our understanding of CTV intraspecific diversity. The elements and conclusions reached in the previous EFSA CTV opinion (EFSA PLH Panel, 2014) are still current and are therefore provided, as a citation of the 2014 opinion (between quotation marks and in italics), in what follows.
2.1.2. Database search
Pest information, on host(s) and distribution, was retrieved from the EPPO Global Database (EPPO, 2017).
Data about import of commodity types that could potentially provide a pathway for the pest to enter the EU and about the area of hosts grown in the EU were obtained from EUROSTAT.
The Europhyt database was consulted for pest‐specific notifications on interceptions and outbreaks. Europhyt is a web‐based network launched by the Directorate General for Health and Consumers (DG SANCO), and is a subproject of PHYSAN (Phyto‐Sanitary Controls) specifically concerned with plant health information. The Europhyt database manages notifications of interceptions of plants or plant products that do not comply with EU legislation, as well as notifications of plant pests detected in the territory of the MSs and the phytosanitary measures taken to eradicate or avoid their spread.
The NCBI GenBank database was consulted to obtain information on partial and complete genomic CTV sequences on August 25, 2017 (NCBI, 2017).
2.2. Methodologies
The Panel performed the pest categorisation for CTV (non‐European isolates), following guiding principles and steps presented in the EFSA guidance on the harmonised framework for pest risk assessment (EFSA PLH Panel, 2010) and as defined in the International Standard for Phytosanitary Measures No 11 (FAO, 2013) and No 21 (FAO, 2004).
In accordance with the guidance on a harmonised framework for pest risk assessment in the EU (EFSA PLH Panel, 2010), this work was initiated following an evaluation of the EU's plant health regime. Therefore, to facilitate the decision‐making process, in the conclusions of the pest categorisation, the Panel addresses explicitly each criterion for a Union quarantine pest and for a Union RNQP in accordance with Regulation (EU) 2016/2031 on protective measures against pests of plants, and includes additional information required as per the specific terms of reference received by the European Commission. In addition, for each conclusion, the Panel provides a short description of its associated uncertainty.
Table 1 presents the Regulation (EU) 2016/2031 pest categorisation criteria on which the Panel bases its conclusions. All relevant criteria have to be met for the pest to potentially qualify either as a quarantine pest or as a RNQP. If one of the criteria is not met, the pest will not qualify. Note that a pest that does not qualify as a quarantine pest may still qualify as a RNQP which needs to be addressed in the opinion. For the pests regulated in the protected zones only, the scope of the categorisation is the territory of the protected zone, thus the criteria refer to the protected zone instead of the EU territory.
Table 1.
Pest categorisation criteria under evaluation, as defined in Regulation (EU) 2016/2031 on protective measures against pests of plants (the number of the relevant sections of the pest categorisation is shown in brackets in the first column)
| Criterion of pest categorisation | Criterion in Regulation (EU) 2016/2031 regarding Union quarantine pest | Criterion in Regulation (EU) 2016/2031 regarding protected zone quarantine pest (articles 32–35) | Criterion in Regulation (EU) 2016/2031 regarding Union regulated non‐quarantine pest |
|---|---|---|---|
| Identity of the pest (Section 3.1 ) | Is the identity of the pest established, or has it been shown to produce consistent symptoms and to be transmissible? | Is the identity of the pest established, or has it been shown to produce consistent symptoms and to be transmissible? | Is the identity of the pest established, or has it been shown to produce consistent symptoms and to be transmissible? |
| Absence/presence of the pest in the EU territory (Section 3.2 ) | Is the pest present in the EU territory?If present, is the pest widely distributed within the EU? Describe the pest distribution briefly! | Is the pest present in the EU territory? If not, it cannot be a protected zone quarantine organism. | Is the pest present in the EU territory? If not, it cannot be a regulated non‐quarantine pest. (A regulated non‐quarantine pest must be present in the risk assessment area). |
| Regulatory status (Section 3.3 ) | If the pest is present in the EU but not widely distributed in the risk assessment area, it should be under official control or expected to be under official control in the near future. | The protected zone system aligns with the pest free area system under the International Plant Protection Convention (IPPC).The pest satisfies the IPPC definition of a quarantine pest that is not present in the risk assessment area (i.e. protected zone). | Is the pest regulated as a quarantine pest? If currently regulated as a quarantine pest, are there grounds to consider its status could be revoked? |
| Pest potential for entry, establishment and spread in the EU territory (Section 3.4 ) | Is the pest able to enter into, become established in, and spread within, the EU territory? If yes, briefly list the pathways! | Is the pest able to enter into, become established in, and spread within, the protected zone areas?Is entry by natural spread from EU areas where the pest is present possible? | Is spread mainly via specific plants for planting, rather than via natural spread or via movement of plant products or other objects?Clearly state if plants for planting is the main pathway! |
| Potential for consequences in the EU territory (section 3.5 ) | Would the pests’ introduction have an economic or environmental impact on the EU territory? | Would the pests’ introduction have an economic or environmental impact on the protected zone areas? | Does the presence of the pest on plants for planting have an economic impact, as regards the intended use of those plants for planting? |
| Available measures (Section 3.6 ) | Are there measures available to prevent the entry into, establishment within or spread of the pest within the EU such that the risk becomes mitigated? |
Are there measures available to prevent the entry into, establishment within or spread of the pest within the protected zone areas such that the risk becomes mitigated? Is it possible to eradicate the pest in a restricted area within 24 months (or a period longer than 24 months where the biology of the organism so justifies) after the presence of the pest was confirmed in the protected zone? |
Are there measures available to prevent pest presence on plants for planting such that the risk becomes mitigated? |
| Conclusion of pest categorisation (Section 4 ) | A statement as to whether (1) all criteria assessed by EFSA above for consideration as a potential quarantine pest were met and (2) if not, which one(s) were not met. | A statement as to whether (1) all criteria assessed by EFSA above for consideration as potential protected zone quarantine pest were met, and (2) if not, which one(s) were not met. | A statement as to whether (1) all criteria assessed by EFSA above for consideration as a potential regulated non‐quarantine pest were met, and (2) if not, which one(s) were not met. |
It should be noted that the Panel's conclusions are formulated respecting its remit and particularly with regards to the principle of separation between risk assessment and risk management (EFSA founding regulation (EU) No 178/2002); therefore, instead of determining whether the pest is likely to have an unacceptable impact, the Panel will present a summary of the observed pest impacts. Economic impacts are expressed in terms of yield and quality losses and not in monetary terms, while addressing social impacts is outside the remit of the Panel, in agreement with EFSA guidance on a harmonised framework for pest risk assessment (EFSA PLH Panel, 2010).
The Panel will not indicate in its conclusions of the pest categorisation whether to continue the risk assessment process, but, following the agreed two‐step approach, will continue only if requested by the risk managers. However, during the categorisation process, experts may identify key elements and knowledge gaps that could contribute significant uncertainty to a future assessment of risk. It would be useful to identify and highlight such gaps so that potential future requests can specifically target the major elements of uncertainty, perhaps suggesting specific scenarios to examine.
3. Pest categorisation
3.1. Identity and biology of the pest
3.1.1. Identity and taxonomy
Is the identity of the pest established, or has it been shown to produce consistent symptoms and to be transmissible? (Yes or N)
YES
Citrus tristeza virus is a well characterised virus in the genus Closterovirus of the Closteroviridae family (Karasev and Bar‐Joseph, 2010; Martelli et al., 2012). It has a large, ca. 19 kilobases positive sense, single‐stranded RNA genome and complete or partial genomic sequences are available for a large number of CTV isolates (EFSA PLH Panel, 2014).
3.1.2. Biology of the pest
The biological properties described apply to all CTV isolates, and there is no information to suggest that non‐EU CTV isolates differ from EU ones in these respects. However, different CTV isolates can cause considerably different symptoms in citrus and can differ in their vector transmission properties.
CTV is a phloem‐associated virus. It replicates in the cytoplasm of companion or phloem parenchyma cells of its hosts. It is therefore graft‐transmissible agent which, as other plant viruses, is transmitted through the vegetative multiplication of infected host plants. In addition, similar to other closteroviruses, it is transmitted by aphids in a semi‐persistent manner (Yokomi et al., 1989). It is not known to be seed‐ (McClean, 1957) or pollen‐transmitted in any of its hosts (Moreno et al., 2008).
CTV is transmitted by several aphid species (Michaud, 1998; Moreno et al., 2008). Five minutes to a few hours of feeding are sufficient for virus acquisition. There is no latency period and the aphids remain viruliferous for only about 24 h, infectivity being completely lost within 48 hours of virus acquisition (Raccah et al., 1976). Toxoptera citricida (Kirkaldy) is the most efficient vector of CTV (Michaud, 1998; Moreno et al., 2008; Gottwald, 2010). Aphis gossypii (Glover), although somewhat less efficient than T. citricida, is also an effective vector (Yokomi et al., 1994). Under experimental conditions Aphis spiraecola (Patch, formerly A. citricola van der Goot) and Toxoptera aurantii (Boyer de Fonscolombe) are able to transmit CTV (Hermoso de Mendoza et al., 1984; Yokomi and Garnsey, 1987). They are however considered to be less efficient and less important vectors than the two other species. Transmission efficiency also varies between CTV isolates.
The known natural host range of CTV is restricted to species of the genera Citrus, Poncirus and Fortunella (subfamily Aurantioidae, family Rutaceae, Moreno et al., 2008). Depending on host species, cultivar and CTV isolate, CTV may cause a variety of symptoms in these hosts.
3.1.3. Intraspecific diversity
During categorisation of EU CTV isolates (EFSA PLH Panel, 2014), the Plant Health panel conducted an extensive analysis of the information available on the intraspecific molecular and biological diversity of CTV. Although many new genomic sequences of CTV isolates have been reported since then,4 these sequences do not modify in any major way, our understanding of CTV intraspecific diversity. The elements and conclusions reached in the previous EFSA CTV opinion (EFSA PLH Panel, 2014) are still current and are therefore provided, as a citation of the 2014 opinion (between quotation marks and in italics), in what follows.
3.1.3.1. Serological and molecular diversity
‘There is ample evidence for serological diversity, and monoclonal antibodies have been generated that react against either a broad spectrum of CTV isolates or with very specific isolates. The antibody MCA13 reacts only with severe CTV isolates (Permar et al., 1989) and is used to discriminate between mild (non‐decline‐ and non‐stem pitting disease (SP)‐inducing) and severe (decline‐ or SP‐inducing) isolates. The molecular diversity of CTV was evident from analyses of partial genome sequences (Ayllón et al., 2001), but when a comprehensive dataset of full genome sequences became available, a more complete definition of CTV strains was possible. Following the most recent review of current knowledge on CTV, virus isolates of this species have been grouped into strains (Harper, 2013)’.
‘It should however be noted that the term “strain” has been very loosely used in the literature in the past, sometimes as a synonym for “isolate” and sometimes to regroup isolates based on their biological properties, or on a combination of the molecular and predicted biological properties. As a consequence of this loose and inconsistent use of terminology, the literature is frequently confusing’.
‘Because recombination was shown to have contributed significantly to the evolutionary history of some isolates or strains of CTV (Vives et al., 2005; Melzer et al., 2010; Harper, 2013), the entire genome sequence is currently taken into account for the taxonomic assignation of isolates to CTV strains. For strain demarcation, the complete genome sequence has to differ by >7.5% (and the sequence of either ORF1a or the encoded protein by >8%). Recombination analyses of representatives of the recognised strains are also required (Harper, 2013). However, for practical reasons, assignation of an isolate to a particular strain has been (and often still is) frequently based on short genome sequence fragments obtained following polymerase chain reaction (PCR) amplification’.
3.1.3.2. Biological diversity
‘Three major syndromes are associated with CTV infections in citrus: tristeza, SP5 and seedling yellows (SY, Moreno and Garnsey, 2010; Dawson et al., 2013). Tristeza is a decline syndrome caused by the vast majority of CTV isolates in different citrus species such as sweet orange (Citrus sinensis), mandarins (C. reticulata), grapefruits (C. paradisi Macfadyen), kumquats (Fortunella sp.) and limes (C. aurantifolia (Christm.) Swingle) when grafted on rootstocks of sour orange (C. aurantium) or lemon (C. limon). Tristeza is therefore a bud union disease that develops only in susceptible rootstocks/scion combinations. The observed decline can be extremely rapid (‘’quick decline”), with wilting and death of trees occurring within a few days or weeks, or it can be a slower process, occurring over months or even years’.
‘SP is the second type of syndrome associated with CTV infection. It occurs in susceptible species regardless of the rootstock used, and can affect both rootstock and grafted varieties (Moreno et al., 2008). It is characterised by the development of pits in the trunk and stem resulting from cambium malfunctioning. SP symptoms are associated with decreased tree vigour, dwarfing of plants and reduced fruit yield and quality’.
‘SY is a CTV‐induced syndrome observed in young plants, most notably under greenhouse conditions. It is characterised by a general yellowing and stunting of affected seedlings and is mostly observed in sour orange, lemons and grapefruit (Moreno et al., 2008)’.
‘There is biological variability in the ability of CTV isolates to cause these three types of syndromes in susceptible hosts (Moreno et al., 2008) and, consequently, CTV isolates have been grouped into pathogenic categories (Garnsey et al., 2005). Within the limits of the assays, symptom differences can be attributed to properties of the infecting CTV isolate. When sour orange is used as a rootstock, the majority of CTV isolates are able to cause tristeza decline symptoms; however, some isolates, such as the T385 Spanish isolate, do not appear to cause decline and are therefore often referred to as “mild isolates” (Vives et al., 1999; Moreno et al., 2008). This term is also commonly used to refer to isolates unable to cause SP or SY symptoms, adding confusion to the literature. Similarly, the term “severe isolates” is used to describe decline‐inducing isolates (in particular in quick decline situations) but, confusingly, is also used to describe isolates causing SP or SY’.
‘CTV isolates also show variability in their ability to overcome the CTV resistance observed in trifoliate orange (Poncirus trifoliata). P. trifoliata is used as a rootstock, albeit not extensively, in Europe. While the majority of virus isolates cannot infect trifoliate orange, a few recombinant RB isolates have been described (Harper et al., 2010) that can overcome this resistance, and are able to replicate in and systemically invade resistant plants’.
3.1.3.3. Correlation between molecular and biological diversity
‘By combining host response, serological and molecular data, efforts were made to establish clear and reproducible correlations between molecular variability of virus isolates/strains and their biological (pathogenic) properties. Genome sequences of reference isolates with experimentally well‐characterised pathogenicities (mild isolate T30 from Florida, severe isolate T36 from Florida [decline‐ and SY‐inducing), SP‐inducing isolates T3 and VT from Florida and Israel (Garnsey et al., 2005)] were determined. This provided a framework of CTV reference isolates to which sequences, biological properties and virulence of newly characterised isolates could be compared. This showed that, to a certain extent, biological properties correlated with those of the most closely related reference (Moreno et al., 2008; Roy and Brlansky, 2009)’.
‘However, growing evidence from sequencing and biological assays demonstrates that CTV isolates assigned to a particular strain can differ remarkably in their abilities to induce particular symptoms; therefore, the notion of a tight correlation between CTV strains and the symptoms induced is no longer valid (Harper, 2013). As with other viruses, slight differences in sequence can lead to important changes in the phenotype of the disease induced (Moreno et al., 2008; Harper, 2013);6 as a result, CTV strains cannot be considered to be a homogenous ensemble of isolates sharing identical pathogenicity profiles. Similarly, the monoclonal antibody CTV MCA13 (Permar et al., 1989), commonly used to identify severe (tristeza‐ and SP‐inducing) isolates, can sometimes react with mild isolates (Hilf and Garnsey, 2002), which, as shown by complete genome sequencing (Varveri et al., 2014), is probably caused by mutations in the region where the neotope for MCA13 is localised’.
‘The analysis of CTV infections in citrus has also revealed that, as with other RNA viruses, infected plants may contain a pool of sequence variants that may belong to a single strain or even to several strains (Rubio et al., 2001). Thus, CTV isolates often comprise mixed virus populations (Harper, 2013), further complicating the analysis of the symptoms caused by individual variants/strains. There is essentially no understanding of how combinations of virus genotypes affect disease symptoms and severity, further complicating any efforts to establish a connection between virus genotype and disease phenotype (Harper, 2013)’. ‘Unfortunately, much confusion in the literature has resulted from initial attempts to ascribe specific pathogenic properties to CTV strains and, later, from attempts to dispell the underlying hypothesis’.
3.1.3.4. Diversity of European CTV isolates
‘Partial or complete genome sequences of a number of European CTV isolates are available, and these demonstrate the presence of several CTV strains (Rubio et al., 2001). Several CTV isolates/strains (e.g. RB isolates) are not known to occur in Europe. From a biological perspective, both tristeza decline‐inducing isolates and mild isolates, unable to induce decline in susceptible rootstock/scion combinations, are known in Europe (Varveri et al., 2014). CTV isolates causing severe SY symptoms in citrus have also been reported (Ferretti et al., 2014). Although sequence variants genetically similar to those of the SP‐inducing non‐European CTV isolates have been detected in the EU (Ruiz‐Ruiz et al., 2006), and have even been implicated in outbreaks with severe tristeza decline symptoms (Owen et al., 2014), SP symptoms in sweet orange have not been observed in field surveys and only rarely occurring, inconspicuous symptoms were induced in indicator plants in the greenhouse (Ballester‐Olmos et al., 1993; Pedro Moreno, Valencian Institute for Agricultural Research, personal communication, 2014). RB isolates which can overcome P. trifoliata resistance have been found in New Zealand (Harper et al., 2010), and sequence variants similar to those of the RB isolates have been reported in a few additional countries outside of Europe but not in the EU (Mariano Cambra, Valencian Institute for Agricultural Research, personal communication, 2014)’.7
‘Overall, European CTV isolates appear to represent only a fraction of the biological and molecular diversity present in CTV isolates throughout the world. Given that, aside from the pathogenic properties of virus isolates characterised on a limited set of indicator hosts, the biological properties of European CTV populations are incompletely understood, this general evaluation is associated with significant uncertainties’.
Conversely, it is clear that non‐EU CTV isolates present a broader molecular and genetic diversity than EU‐ones. In particular, non‐EU isolates comprise some isolates able to cause severe SP symptoms in sweet orange, or RB isolates that can overcome the resistance of trifoliate orange and its hybrids (Harper et al., 2010), all of which do not appear to have so far equivalents in populations of EU isolates. This conclusion is however associated with significant uncertainties, in particular because the diversity of CTV isolates present in ornamental citrus species such as kumquats or calamondin has been very little analysed to date.
3.1.4. Detection and identification of the pest
Are detection and identification methods available for the pest?
YES, for CTV in general but there are no specific assays for the detection of non‐EU isolates
As extensively described in the previous EFSA opinion on CTV (EFSA PLH Panel, 2014) a wide range of techniques are available for the detection of CTV, including graft‐inoculation of indicator plants (Wallace and Drake, 1951; Garnsey et al., 2005; Pina et al., 2005), serological assays (Garnsey and Cambra, 1991; Garnsey et al., 1993; Cambra et al., 2000a,b), molecular tests based on reverse transcription polymerase chain reaction (RT‐PCR) (Nolasco et al., 1993; Olmos et al., 1996) or real‐time PCR (Bertolini et al., 2008; Vidal et al., 2012). Real‐time PCR with genotype‐specific probes (Ruiz Ruiz et al., 2009; Ananthakrishnan et al., 2010) or genotyping using a miniaturised silicon lab‐on‐chip (LoC) device (Scuderi et al., 2016) allow the specific detection and quantification of CTV strains8 even in mixed infection. Standard protocols allowing the unequivocal identification of CTV are available (EPPO, 2004). With the availability of high‐throughput sequencing methods complete genome sequence determination is also more and more used for CTV isolates characterisation.
However, as stated in the previous EFSA Opinion on CTV (EFSA PLH Panel, 2014) ‘in the absence of appropriate biological assays (Garnsey et al., 2005; Wang et al., 2013), these methods appear of limited value for the prediction of pathogenic properties of CTV isolates (Bar‐Joseph et al., 2010; Harper et al., 2010). Therefore, a combination of biological, molecular and, possibly, serological data are needed for a conclusive characterisation of the genetic and pathogenic features of a CTV isolate’.
3.2. Pest distribution
3.2.1. Pest distribution outside the EU
As indicated in the previous EFSA Opinion on CTV (EFSA PLH Panel, 2014) ‘CTV is originally a pathogen of non‐European origin [and] has been recorded in most citrus‐growing areas of all five continents. In general, country reports do not specify the presence of particular CTV isolates/strains or of the biological properties of the isolates; however, RB isolates have been specifically reported from New Zealand (Harper et al., 2010) and, more recently, from Puerto Rico, where they have most likely been present since 1992 (Roy et al., 2013). In addition, outside of Europe, in the main citrus‐producing countries of the world, CTV isolates causing SP appear to be present and prevalent, and in some citrus‐producing industries cross‐protection against these CTV isolates is necessary for economic production (Moreno et al., 2008)’ (Figure 1).
Figure 1.
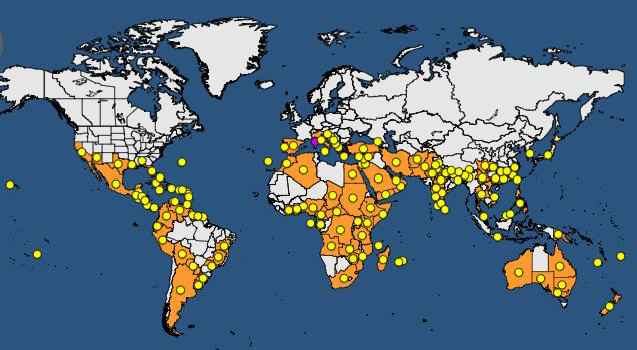
Global distribution of Citrus tristeza virus (non‐EU isolates) (extracted from EPPO Global Database, accessed 28 September 2017)
Last updated: 2017‐09‐13
3.2.2. Pest distribution in the EU
Is the pest present in the EU territory? If present, is the pest widely distributed within the EU?
NO, non‐EU isolates of CTV are not known to be present in the EU
As indicated in the previous EFSA Opinion on CTV (EFSA PLH Panel, 2014) ‘Based on MSs’ answers to the EFSA questionnaire, CTV is present in seven out of the eight EU MSs [Spain, Italy, Greece, Cyprus, Croatia, Portugal and France] with significant citrus production (according to the Eurostat database, see Table 8). In Malta, where virus surveys are continuously conducted (Attard et al., 2009), occasional findings of CTV have been followed by eradication efforts, and CTV is now considered to be eradicated there (Table 2). For other MSs, CTV is considered transient, under eradication (France), present with few occurrences (Greece) or with restricted distribution (Cyprus, Italy), or present but with parts of the country still unaffected (Portugal). CTV is present and widespread in Spain and Croatia.9 With regards to France, the protected zone status of Corsica has recently been removed (Commission Implementing Directive 214/78/EU). The most recent reports of CTV interception are from Italy, France and Portugal and concern CTV found in sweet orange (C. sinensis) and mandarin (C. reticulata) plants imported from Spain’.
Table 2.
Distribution outside the EU of Citrus tristeza virus (extracted from EPPO Global Database, accessed 28 September 2017)
| Continent | Country |
|---|---|
| Africa | Algeria, Angola; Benin; Cameron; Central African Republic; Chad; Comoros; Democratic republic of the Congo; Cote d'Ivoire; Egypt; Ethiopia; Gabon; Ghana; Kenya; Libya; Madagascar; Mauritius; Morocco; Mozambique, Nigeria, Reunion; Sao Tome & Principe; Somalia; South Africa; Sudan; Tanzania; Tunisia; Uganda; Zambia; Zimbabwe; |
| America | Antigua and Barbuda; Argentina; Aruba; Bahamas; Belize; Bermuda; Bolivia; Brasil; Chile; Columbia; Costa Rica; Cuba; Dominica; Dominican Republic; Ecuador; El Salvador; French Guiana; Guadeloupe; Guatemala; Guyana; Honduras; Jamaica; Martinique; Mexico; Netherlands Antilles; Nicaragua, Panama; Paraguay; Peru; Puerto Rico; Saint Lucia; Suriname; Trinidad and Tobago; United States of America; Uruguay; Venezuela; Virgin Islands (British) |
| Asia | Afghanistan; Brunei Darussalam; China; India; Indonesia; Iran; Israel; Japan; Jordan; Korea, Republic; Lebanon; Malaysia, Nepal; Oman; Pakistan; Philippines; Saudi Arabia; Sri Lanka; Syria; Taiwan; Thailand; United Arab Emirates; Vietnam; Yemen; |
| Europe | Albania; Bosnia and Herzegovina; Georgia; Montenegro; Turkey |
| Oceania | American Samoa; Australia; Fiji; French Polynesia; New Caledonia; New Zealand; Papua New Guinea; Samoa; Tonga |
‘In general, CTV infections in Europe in citrus species grafted on sour orange rootstocks are characterised by typical tristeza rapid decline symptoms, ranging in severity, or by no symptoms at all (Ballester‐Olmos et al., 1993; Moreno et al., 2008), the latter situation corresponding to mild isolates unable to cause decline (Varveri et al., 2014). Irrespective of the rootstock/scion combination, symptoms of SP have not yet been observed on sweet orange in the field in Europe.10 Despite this, CTV genotypes closely related to isolates found in other parts of the world, and associated with severe SP symptoms, have been reported in Sicily (Davino et al., 2005; Rizza et al., 2007),11 Spain (Ruiz‐Ruiz et al., 2006), Crete (Owen et al., 2014), Greece (Malandraki et al., 2011) and the east Adriatic region (mainly Croatia and Montenegro, Cerni et al., 2009). CTV genotypes representing the RB strain, able to “break” the resistance of P. trifoliata, are not known to occur Europe’.
So, overall, the Panel concludes that non‐EU isolates of CTV are not present in the EU and therefore do not meet this criterion to qualify as a Union RNQP. However, as stated in the 2014 CTV Opinion, there are uncertainties attached to this evaluation. In particular ‘There are uncertainties about the reason(s) for the apparent inability of CTV isolates, closely related to SP‐inducing isolates, to cause SP symptoms in sweet orange orchards in Europe, and about the potential mid‐ and long‐term evolution of this situation’. And ‘Another area of uncertainty concerns the extremely limited information available on the prevalence and biological properties of CTV isolates that may be present in ornamental citrus such as kumquats (Fortunella sp.) and calamondin (Citrofortunella microcarpa) in Europe’.
3.2.3. Vectors and their distribution in the EU
T. citricida is the most efficient vector of CTV. It is a regulated pest listed in Annex IIAI of Council Directive 2000/29/EC. In the EU, it is reported only from Portugal and Spain, in both cases with a restricted distribution (EPPO GD accessed on 29 June 2017) and away from the most important citrus‐producing areas of these countries. It is however reported by the same source as widespread in Madeira.
A. gossypii, A. spiraecola and T. aurantii, the other known CTV vector species, are present in Europe. In particular, A. gossypii, the second most efficient vector, is widespread in the EU (Figure 2).
Figure 2.
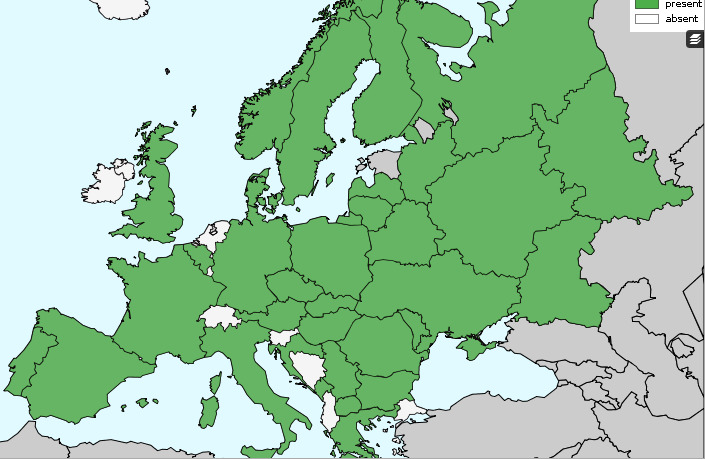
Current distribution of the Aphis gossypii vector of Citrus tristeza virus extracted from Fauna Europea Database accessed August 28, 2017 (de Jong et al., 2014)
As stated in the previous EFSA Opinion on CTV (EFSA PLH Panel, 2014 ) ‘The efficiency by which CTV isolates are transmitted by A. gossypii varies with the particular virus isolate, but is generally greater than 50% and thus, with its high population sizes, A. gossypii plays a major role in epidemics of CTV in Spain (Cambra et al., 2000a) and across Europe. Overall, and with minimal uncertainty, aphid vectors, with the potential to contribute to CTV spread, can be considered to be widely available in the EU’.
3.3. Regulatory status
3.3.1. Council Directive 2000/29/EC
Citrus tristeza virus (non‐European isolates) is listed in Council Directive 2000/29/EC. Details are presented in Tables 3 and 4.
Table 3.
Citrus tristeza virus (non‐European isolates) in Council Directive 2000/29/EC
| Annex II, Part A | Harmful organisms whose introduction into, and spread within, all member states shall be banned if they are present on certain plants or plant products | |
| Section I | Harmful organisms not known to occur in the community and relevant for the entire community | |
| (d) | Virus and virus‐like organisms | |
| Species | Subject of contamination | |
| 7 | Citrus tristeza virus (non‐European isolates) | Plants of Citrus L., Fortunella Swingle, Poncirus Raf., and their hybrids, other than fruit and seeds |
3.3.2. Legislation addressing plants and plant parts on which Citrus tristeza virus (non‐European isolates) is regulated
Table 4.
Regulated hosts and commodities that may involve Citrus tristeza virus (non‐European isolates) in Annexes III, IV and V of Council Directive 2000/29/EC
| Annex III, Part A | Plants, plant products and other objects the introduction of which shall be prohibited in all member states |
|---|---|
| Description | Country of origin |
| 16. Plants of Citrus L., Fortunella Swinlge, Poncirus Raf., and their hybrids, other than fruit and seeds | Third countries |
| Annex IV, Part A | Special requirements which must be laid down by all member states for the introduction and movement of plants, plant products and other objects into and within all member states |
| Section I | Plants, plant products and other objects originating outside the community |
| Plants, plant products and other objects | Special requirements |
| 16.1 Fruits of Citrus L., Fortunella Swingle, Poncirus Raf., and their hybrids, originating in third countries | The fruits shall be free from peduncles and leaves and the packaging shall bear an appropriate origin mark. |
| Section II | Plants, plant products and other objects originating in the community |
| Plants, plant products and other objects | Special requirements |
| 30.1 Fruits of Citrus L., Fortunella Swingle, Poncirus Raf., and their hybrids | The packaging shall bear an appropriate origin mark |
| Annex V | Plants, plant products and other objects which must be subject to a plant health inspection (at the place of production if originating in the community, before being moved within the community — in the country of origin or the consignor country, if originating outside the community) before being permitted to enter the community |
| Part A |
Plants, plant products and other objects originating in the community I.Plants, plant products and other objects which are potential carriers of harmful organisms of relevance for the entire Community and which must be accompanied by a plant passport |
|
1.4 Plants of Fortunella Swingle, Poncirus Raf., and their hybrids, Casimiroa La Llave, Clausena Burm. f., Vepris Comm., Zanthoxylum L. and Vitis L., other than fruit and seeds. 1.5 Without prejudice to point 1.6, plants of Citrus L. and their hybrids other than fruit and seeds. 1.6 Fruits of Citrus L., Fortunella Swingle, Poncirus Raf. and their hybrids with leaves and peduncles. |
|
| Part B |
Plants, plant products and other objects originating in territories, other than those territories referred to in part A. I. Plants, plant products and other objects which are potential carriers of harmful organisms of relevance for the entire Community |
|
1. Plants, intended for planting, other than seeds but including seeds of ….. Citrus L., Fortunella Swingle and Poncirus Raf., and their hybrids…… 3. Fruits of: ‐ Citrus L., Fortunella Swingle, Poncirus Raf., and their hybrids, Momordica L. and Solanum melongena L. |
3.3.3. Legislation addressing vectors of Citrus tristeza virus (non‐European isolates)
Table 5.
Toxoptera citricida in Council Directive 2000/29/EC
| Annex II, Part A | Harmful organisms whose introduction into, and whose spread within, all Member States shall be banned if they are present on certain plants or plant products, | |
| Section I | Harmful organisms not known to occur in the Community and relevant for the entire Community, | |
| (a) | Insects, mites and nematodes, at all stages of their development | |
| Species | Subject of contamination | |
| 30. | Toxoptera citricida | Plants of Citrus L., Fortunella Swingle, Poncirus Raf., and their hybrids, other than fruit and seeds |
3.3.4. Marketing directive
Host plants of CTV are explicitly mentioned in the Council Directive 2008/90/EC12.
3.4. Entry, establishment and spread in the EU
3.4.1. Host range
As stated in the previous EFSA Opinion on CTV (EFSA PLH Panel, 2014), ‘CTV has a restricted host range, and plants of Citrus spp., including lemon, lime, sweet and sour orange, tangerine, mandarin, grapefruit; Fortunella spp., a genus comprising several kumquat species (Moreno et al., 2008); and Poncirus spp. are the only known natural hosts. Citrus species are widely cultivated in the Mediterranean part of the EU, while kumquats and some other citrus species, such as calamondin, are cultivated mainly as ornamental trees and have a more limited commercial importance’.
In addition ‘Several plant species belonging to other genera within the subfamily Aurantioideae (Aegle, Aeglopsis, Afraegle, Atalantia, Citropsis, Clausena, Eremocitrus, Hesperethusa, Merrillia, Microcitrus, Pamburus, Pleiospermium and Swinglea) have been shown to be experimental hosts of CTV (Moreno et al., 2008). CTV has also been experimentally transmitted to Passiflora gracilis and P. caerulea [family Passifloraceae (Kitajima et al., 1974; Müller et al., 1974; Roistacher and Bar‐Joseph, 1987)]. However, experimental hosts of CTV, outside of the Rutaceae family, are unlikely to have any practical significance. Uncertainties exist on the status of Rutaceae other than Citrus, Fortunella and Poncirus as natural hosts for CTV, especially those that are used as ornamentals, and about their potential significance for virus dissemination and CTV epidemiology’.
3.4.2. Entry
Is the pest able to enter into the EU territory? (Yes or No) If yes, identify and list the pathways!
YES, CTV can enter via trade of non‐regulated host plants
The most important pathway for entry, the trade of plants for planting of the known host species of CTV, Citrus, Fortunella and Poncirus and their hybrids is closed by the existing Annex III legislation (see 3.3.2 and Table 4 above). As a consequence, entry is only considered to be possible on alternative, low probability and/or high uncertainty pathways:
Trade of plants of Rutaceae species which are not known to be natural hosts of CTV but have been shown to be experimental hosts (see Section 3.4.1).
Entry of viruliferous vectors on unregulated plants or plant products or as hitchhikers, but infectivity is lost rapidly (see Section 3.1.2) so that the probability of transfer to a suitable host would appear to the very low.
Illegal entry of infected plants for planting of susceptible host species for commercial or for personal use.
Between 1995 and 24 August 2017, there were 21 records of interception of Citrus tristeza virus in the Europhyt database. The database does not separate between interceptions of EU and non‐EU CTV isolates but all 21 interceptions concern intra‐EU trade and therefore presumably concern only EU isolates
3.4.3. Establishment
Is the pest able to become established in the EU territory?
YES, hosts are widely present in the EU
3.4.3.1. EU distribution of main host plants
Citrus sp. hosts of CTV are commercially grown for citrus fruit production (oranges, mandarins, lemons, etc.) in eight Members States of the EU. In order of decreasing production they are: Spain, Italy, Greece, Portugal, Cyprus, Croatia, Malta and France. In addition, plants of Citrus, Fortunella and Poncirus are grown as ornamentals, either in the open or under protected cultivation in a number of Member States (Table 6).
Table 6.
Area (cultivation/harvested/production) of citrus production (in 1,000 ha) in Europe according to the Eurostat database (Crop statistics apro_acs_a, extracted on 20 June 2017)
| GEO/TIME | 2012 | 2013 | 2014 | 2015 | 2016 |
|---|---|---|---|---|---|
| Spain | 310.50 | 306.31 | 302.46 | 298.72 | 295.33 |
| Italy | 146.79 | 163.59 | 140.16 | 149.10 | 141.22 |
| Greece | 50.61 | 49.88 | 49.54 | 46.92 | 44.72 |
| Portugal | 19.85 | 19.82 | 19.80 | 20.21 | 20.21 |
| France | 3.89 | 4.34 | 4.16 | 4.21 | 4.70 |
| Cyprus | 3.21 | 2.63 | 2.69 | 2.84 | 3.29 |
| Croatia | 1.88 | 2.17 | 2.17 | 2.21 | 2.18 |
Last update 14‐6‐17
3.4.3.2. Climatic conditions affecting establishment
As stated in the previous EFSA Opinion on CTV (EFSA PLH Panel, 2014) ‘The ecoclimatic requirements of CTV are similar to those of its host plants and therefore it is not expected to be limited by ecoclimatic conditions in areas where its hosts are able to develop. Citrus cultivation occurs in the warmer regions of Europe, where citrus plants are widely grown in orchards (see EFSA PLH Panel, 2014)’. Indeed isolates of CTV have already established in seven of the eight EU members States were Citrus are commercially grown.
3.4.4. Spread
3.4.4.1. Vectors and their distribution in the EU
Is the pest able to spread within the EU territory following establishment? (Yes or No) How?
YES, though the action of aphid vectors and through the trade of infected plants for planting
RNQPs: Is spread mainly via specific plants for planting, rather than via natural spread or via movement of plant products or other objects?
YES
As stated in the previous EFSA Opinion on CTV (EFSA PLH Panel, 2014), ‘The rate of CTV transmission in the field is influenced by many factors, including the composition and density of aphid populations, environmental conditions and the susceptibility of citrus species and varieties present (Moreno et al., 2008). In Europe, given the restricted presence of the very efficient T. citricida vector, A. gossypii is the most relevant vector for CTV spread, and disease epidemics are associated with this vector (Gottwald et al., 1997; Cambra et al., 2000a; Davino et al., 2005). Recent evidence from virus/vector studies under laboratory conditions highlights the important role played by A. gossypii in CTV disease outbreaks in Calabria (Campolo et al., 2014). Single A. gossypii insects acquired local CTV isolates after a 30‐min feeding acquisition period and transmitted the virus, in a semi‐persistent transmission mode, after a 60‐min feeding transmission period (Campolo et al., 2014). Only four aphids per plant were needed to reach a 50% CTV transmission probability, thereby demonstrating the ability of local A. gossypii populations to efficiently spread CTV’.
‘Recent studies conducted in various countries (Gottwald et al., 1995; Cambra et al., 2000a; Davino et al., 2005, 2013; Ferretti et al., 2014; Owen et al., 2014) show that spread of CTV in orchards can be rapid […]. Spread is associated with aphid vectors, but also with the movement of vegetatively propagated plants for planting, including ornamental citrus such as calamondin and kumquats (Chatzivassiliou and Nolasco, 2014)’.
Also, ‘Despite a limited number of interception reports (Europhyt database) linking intra‐EU trade of plants for planting with CTV movement, existing citrus certification systems constitute a strong limitation to the CTV spread through the plants for planting pathway’.
Overall, there is very limited uncertainty that if introduced into the EU, non‐EU isolates of CTV would be able to efficiently spread, in a similar fashion and through the same mechanisms that have ensured the spreading of EU isolates.
3.5. Impacts
Would the pests’ introduction have an economic or environmental impact on the EU territory?
YES
RNQPs: Does the presence of the pest on plants for planting have an economic impact, as regards the intended use of those plants for planting? 13
YES
The analysis of potential CTV impacts performed in the frame of the previous EFSA opinion (EFSA PLH Panel, 2014) is still current and is therefore provided here.
‘CTV causes two very serious diseases of citrus, tristeza decline and SP, and has had a serious impact in all major citrus‐growing regions of the world. Almost 100 million trees grafted on susceptible rootstocks have died worldwide from tristeza decline, the affected species being mainly sweet orange (C. sinensis) and mandarin (C. reticulata) (Bar‐Joseph et al., 1989). Affected trees commonly show decline symptoms including foliage yellowing and shedding, twig dieback, progressive reduction of root systems, size decrease and discoloration of fruits, which are eventually followed by plant death. In its most dramatic manifestation, citrus tristeza disease causes a quick decline characterised by the sudden appearance of rapidly progressing symptoms eventually resulting in collapse and death of the tree within days or weeks from symptom onset. Tristeza decline can also be slow, which results in plant deterioration over longer periods of up to several years, sometimes with a latency period of up to 20 years, during which time CTV infection causes only mild symptoms or no symptoms at all (Garnsey and Lee, 1988)’.
‘In contrast to tristeza decline, SP affects mostly lime, grapefruit, and sweet orange (C. sinensis (L.) Osbeck), regardless of the rootstock on which these species are grafted. Symptoms of SP consist of irregular radial growth of the tree or its stems caused by the disruption of meristematic activity at localised parts of the cambium. This generates depressions in the wood that may assume a ropy, channelled, porous or spongy appearance. SP can be accompanied by stunting, yellowing and size reduction of leaves. It affects tree vigour and is associated with a considerable reduction in fruit yield and quality (Bar‐Joseph and Dawson, 2008; Moreno and Garnsey, 2010). However, there is no deterioration or death of affected trees. Despite the fact that European isolates closely related to non‐ European, SP‐inducing isolates have been detected in several EU MSs, SP symptoms have not been observed in sweet orange groves of the EU. There is uncertainty regarding the reasons underlying this observation and concerning possible future developments’.
‘SY consists of stunting, small, pale or yellow leaves, and reduced root systems appearing in sour orange, grapefruit or lemon seedlings. The syndrome is sometimes transitory and followed by recovery of affected plants, which may resume normal growth. SY is generally not considered a major constraint and is mostly observed in greenhouse‐grown plants (Moreno et al., 2008)’.
As non‐EU isolates of CTV are not currently present in the EU, there is currently no impact. However, CTV causes very severe diseases of citrus and can have a very considerable impact on the citrus industry. A range of non‐EU CTV isolates are able to cause the severe SP disease, mostly on lime, grapefruit, and sweet orange, a syndrome against which the European orchards are not protected. There are little uncertainties that the introduction and spread of such isolates would have severe detrimental effects on EU citrus crops. The same would apply to the introduction in the EU of RB isolates.
For the same reasons, the presence of EU or non‐EU isolates of CTV on citrus plants for planting very severely affects their intended use, with very limited uncertainty.
3.6. Availability and limits of mitigation measures
Are there measures available to prevent the entry into, establishment within or spread of the pest within the EU such that the risk becomes mitigated?
NO: as the main pathway is already closed by legislation, it is difficult to address the alternative, low probability and/or high uncertainties pathways
RNQPs: Are there measures available to prevent pest presence on plants for planting such that the risk becomes mitigated?
YES: existing citrus certification systems constitute a strong limitation to CTV spread through plants for planting
3.6.1. Biological or technical factors limiting the feasibility and effectiveness of measures to prevent the entry, establishment and spread of the pest
Efficient transmission of the virus by at least one widespread aphid species
Existence of asymptomatic, mild isolates
Possibility of asymptomatic infection in some hosts (latency, lower susceptibility, etc.).
3.6.2. Biological or technical factors limiting the ability to prevent the presence of the pest on plants for planting
Efficient transmission of the virus by at least one widespread aphid species
Existence of asymptomatic, mild isolates
Possibility of asymptomatic infection in some hosts (latency, lower susceptibility, etc.).
3.6.3. Control methods
Use of rootstocks preventing the development of tristeza decline on the scions. This strategy is however not efficient against SP causing isolates or RB isolates
Cross‐protection against SP isolates by pre‐inoculation of trees with mild protecting isolates
Control of aphid vector populations to limit the spread of CTV. But this measure is relatively inefficient, except in nurseries, given the characteristics of the transmission mode
Use of certified planting material, elimination of infected trees to reduce local inoculum.
3.7. Uncertainty
Four main aspects affected by uncertainties have been identified by the Panel:
Uncertainties on the status of Rutaceae species other than Citrus, Fortunella and Poncirus as natural hosts for CTV, and about their potential significance for virus dissemination and CTV epidemiology.
Uncertainties about whether some non‐EU CTV isolates might be present but not detected in the EU.
Uncertainties about the inability of European CTV isolates, apparently related to non‐European SP‐inducing isolates, to cause SP symptoms in sweet orange groves of the EU.
Uncertainties on the prevalence and biological properties of CTV isolates that may be present in ornamental citrus such as kumquats (Fortunella sp.) and calamondin (C. microcarpa) in Europe.
4. Conclusions
CTV causes very severe diseases of citrus. It has had and will further have a very considerable impact on the EU citrus industry. A range of non‐EU CTV isolates are able to cause the SP disease, mostly on lime, grapefruit, and sweet orange, a syndrome against which the European orchards are not protected. Resistance breaking isolates similarly compromise one of the strategies to control CTV. There are little uncertainties that introduction and spread of such isolates would have major detrimental effects on EU citrus crops (Table 7).
Table 7.
The Panel's conclusions on the pest categorisation criteria defined in Regulation (EU) 2016/2031 on protective measures against pests of plants (the number of the relevant sections of the pest categorisation is shown in brackets in the first column)
| Criterion of pest categorisation | Panel's conclusions against criterion in Regulation (EU) 2016/2031 regarding Union quarantine pest | Panel's conclusions against criterion in Regulation (EU) 2016/2031 regarding Union regulated non‐quarantine pest | Key uncertainties |
|---|---|---|---|
| Identity of the pest (Section 3.1 ) | CTV is a well‐known and well characterised agent | CTV is a well‐known and well characterised agent | No uncertainty |
| Absence/presence of the pest in the EU territory (Section 3.2 ) | Non‐EU CTV isolates are not known to occur in the EU | Non‐EU CTV isolates are not known to occur in the EU. Therefore they do not meet this criterion to qualify as a Union RNQP. | Uncertainties about whether some non‐EU CTV isolates might be present but not detected in the EU and on the prevalence and biological properties of CTV isolates that may be present in ornamental citrus |
| Regulatory status (Section 3.3 ) | CTV non‐EU isolates are currently regulated under Directive 2000/29 | CTV non‐EU isolates currently regulated under Directive 2000/29 | No uncertainty |
| Pest potential for entry, establishment and spread in the EU territory (Section 3.4 ) |
Is the pest able to enter into, become established in, and spread within, the EU territory? YES, but for entry only through alternative minor alternative pathways |
Plants for planting constitute the main means of spread over long distances but this pathway is closed for entry by existing legislation | Uncertainties on the status of Rutaceae species other than Citrus, Fortunella and Poncirus as natural hosts for CTV, and about their potential significance for virus dissemination |
| Potential for consequences in the EU territory (Section 3.5 ) | Introduction and spread of non‐EU, SP‐causing or RB CTV isolates would have severe detrimental effects on EU citrus crops | Because of the negative impact of CTV, its presence on plants for planting of host species would have a negative impact on their intended use | Uncertainties about the inability of European CTV isolates, apparently related to non‐European SP‐inducing isolates, to cause SP symptoms in sweet orange groves of the EU |
| Available measures (Section 3.6 ) |
Use of rootstocks preventing the development of tristeza decline on the scions (but not effective against SP causing isolates) Cross‐protection against SP isolates by pre‐inoculation of trees with mild protecting isolates Use of certified planting material, elimination of infected trees to reduce local inoculum |
Certification of planting material of susceptible host species is by far the most efficient control method, because efficient diagnostics are available | Uncertainties on the status of Rutaceae species other than Citrus, Fortunella and Poncirus as natural hosts for CTV, and about their potential significance for virus dissemination |
| Conclusion on pest categorisation (Section 4 ) | Non‐EU CTV isolates meet all the criteria evaluated by EFSA to qualify as a Union quarantine pest. | Non‐EU CTV isolates do not meet the presence on the territory criterion to qualify as a Union RNQP. | |
| Aspects of assessment to focus on/scenarios to address in future if appropriate |
The main knowledge gaps or uncertainties identified concern:
|
||
Abbreviations
- CTV
Citrus tristeza virus
- DG SANCO
Directorate General for Health and Consumers
- EPPO
European and Mediterranean Plant Protection Organization
- EU MS
European Union Member State
- FAO
Food and Agriculture Organization
- IPPC
International Plant Protection Convention
- LoC
lab‐on‐chip
- PLH
EFSA Panel on Plant Health
- RNQP
regulated non‐quarantine pest
- RT‐PCR
reverse transcription polymerase chain reaction
- SP
stem pitting
- SY
seedling yellows
- TFEU
Treaty on the Functioning of the European Union
- ToR
Terms of Reference
Suggested citation: EFSA PLH (EFSA Panel on Plant Health) , Jeger M, Bragard C, Caffier D, Dehnen‐Schmutz K, Gilioli G, Gregoire J‐C, Jaques Miret JA, MacLeod A, Navajas Navarro M, Niere B, Parnell S, Potting R, Rafoss T, Rossi V, Urek G, Van Bruggen A, Van der Werf W, West J, Chatzivassiliou E, Winter S, Catara A, Duran‐Vila N, Hollo G and Candresse T, 2017. Scientific Opinion on the pest categorisation of Citrus tristeza virus (non‐European isolates). EFSA Journal 2017;15(10):5031, 29 pp. 10.2903/j.efsa.2017.5031
Requestor: European Commission
Question number: EFSA‐Q‐2017‐00310
Panel members: Claude Bragard, David Caffier, Thierry Candresse, Elisavet Chatzivassiliou, Katharina Dehnen‐Schmutz, Gianni Gilioli, Jean‐Claude Gregoire, Josep Anton Jaques Miret, Michael Jeger, Alan MacLeod, Maria Navajas Navarro, Björn Niere, Stephen Parnell, Roel Potting, Trond Rafoss, Vittorio Rossi, Gregor Urek, Ariena Van Bruggen, Wopke Van der Werf, Jonathan West and Stephan Winter.
Adopted: 28 September 2017
Reproduction of the images listed below is prohibited and permission must be sought directly from the copyright holder:
Figure 1: © EPPO;
Figure 2: © Fauna Europea, de Jong 2014
Notes
Council Directive 2000/29/EC of 8 May 2000 on protective measures against the introduction into the Community of organisms harmful to plants or plant products and against their spread within the Community. OJ L 169/1, 10.7.2000, p. 1–112.
Regulation (EU) 2016/2031 of the European Parliament of the Council of 26 October 2016 on protective measures against pests of plants. OJ L 317, 23.11.2016, p. 4–104.
Regulation (EC) No 178/2002 of the European Parliament and of the Council of 28 January 2002 laying down the general principles and requirements of food law, establishing the European Food Safety Authority and laying down procedures in matters of food safety. OJ L 31/1, 1.2.2002, p. 1–24.
62 full length genomic sequences available as of 25 August 2017.
Stem pitting (SP).
Posterior to the quoted EFSA opinion (2014a), Russo et al. (2015) have confirmed this by showing that two asymptomatic and cross protective VT isolates differ by only 13 and 14 point mutations (with 5 and 6 of these silent) from a VT‐SY isolate. Similar results have also been obtained by reverse genetics with the T36 isolate by Folimonova (2012, 2013).
Scuderi et al. (2016) have reported the discovery in two alemow (Citrus macrophylla) seedlings in Sicily of a T36‐like isolate with high homology to RB isolates.
Six major strains, typified by isolates T36, T3, T30, T68, VT and RB are currently recognised (Harper, 2013).
As of 28 August 2017, CTV status in Croatia is reported as ‘present, restricted distribution’ and ‘present, few occurrences’ for Portugal in the EPPO Global Database.
However, some CTV isolates inducing SP on branches of field grapefruit and sweet orange have been reported in Cyprus (Papayiannis et al., 2007), but the Panel was unable to identify further information on the current situation of these isolates
In this specific case, SP was observed on Mexican lime and not on sweet orange and grapefruit. Partial sequence analysis suggested symptoms were caused by a SY isolate. More recent results indicate however that the isolate involved is a VT‐Asian subtype strain (Licciardello et al., 2015).
Council Directive 2008/90/EC of 29 September 2008 on the marketing of fruit plant propagating material and fruit plants intended for fruit production. OJ L 267, 8/10/2008, p. 8–22
See Section 2.1 on what falls outside EFSA's remit.
References
- Ananthakrishnan G, Venkataprasanna T, Roy A and Brlansky RH, 2010. Characterization of the mixture of genotypes of a Citrus tristeza virus isolate by reverse transcription‐quantitative real‐time PCR. Journal of Virological Methods, 164, pp. 75–82. [DOI] [PubMed] [Google Scholar]
- Attard D, Gatt M, Agius MA, Muscat A and Leone Ganado C, 2009. Citrus tristeza virus (CTV) survey in the Maltese Islands 1999–2005. In: D'Onghia AM, Djelouah K and Roistacher CN (eds.). Citrus Tristeza Virus and Toxoptera Citricidus: A Serious Threat to the Mediterranean Citrus Industry. Options Méditerranéennes: Série B. Etudes et Recherches. No 65. CIHEAM, Bari. pp. 76–78. [Google Scholar]
- Ayllón MA, López C, Navas‐Castillo J, Garnsey SM, Guerri J, Flores R and Moreno P, 2001. Polymorphism of the 5′ terminal region of Citrus tristeza virus (CTV) RNA: incidence of three sequence types in isolates of different origin and pathogenicity. Archives of Virology, 146, 27–40. [DOI] [PubMed] [Google Scholar]
- Ballester‐Olmos JF, Pina JA, Carbonell EA, Moreno P, Hermoso de Mendoza A, Cambra M and Navarro L, 1993. Biological diversity of citrus tristeza virus (CTV) isolates in Spain. Plant Pathology, 42, 219–229. [Google Scholar]
- Bar‐Joseph M and Dawson WO, 2008. Citrus tristeza virus. In: Mahy BWJ and Van Regenmortel MHV (eds.). Encyclopedia of Virology. Elsevier Ltd., Amsterdam. pp. 520–525. [Google Scholar]
- Bar‐Joseph M, Marcus R and Lee RF, 1989. The continuous challenge of citrus tristeza virus control. Annual Review of Phytopathology, 27, 291–316. [Google Scholar]
- Bar‐Joseph M, Batuman O and Roistacher C, 2010. The history of Citrus tristeza virus—Revisited. In: Karasev AV and Hilf ME (eds.). Citrus Tristeza Virus Complex and Tristeza Diseases. APS Press, St Paul, MN, USA. pp. 3–26. [Google Scholar]
- Bertolini E, Moreno A, Capote N, Olmos A, De Luis A, Vidal E, Pérez‐Panadés J and Cambra M, 2008. Quantitative detection of Citrus tristeza virus in plant tissues and single aphids by real‐time RT‐PCR. European Journal of Plant Pathology, 120, 177–188. [Google Scholar]
- Cambra M, Gorris MT, Marroquín C, Román MP, Olmos A, Martínez MC, Hermoso de Mendoza A, López A and Navarro L, 2000a. Incidence and epidemiology of Citrus tristeza virus in the Valencian Community of Spain. Virus Research, 71, 85–95. [DOI] [PubMed] [Google Scholar]
- Cambra M, Gorris MT, Román MP, Terrada E, Garnsey SM, Camarasa E, Olmos A and Colomer M, 2000b. Routine detection of citrus tristeza virus by direct Immunoprinting‐ELISA method using specific monoclonal and recombinant antibodies. Proceedings of the 14th Conference of the International Organization of Citrus Virologists. IOCV, Riverside, CA, USA, pp. 34–41.
- Campolo O, Chiera E, Malacrinò A, Laudani F, Fontana A, Albanese GR and Palmeri V, 2014. Acquisition and transmission of selected CTV isolates by Aphis gossypii. Journal of Asia‐Pacific Entomology, 17, 493–498. [Google Scholar]
- Cerni S, Skoric D, Ruscic J, Krajacic M, Papic T, Djelouah K and Nolasco G, 2009. East Adriatic—a reservoir region of severe Citrus tristeza virus strains. European Journal of Plant Pathology, 124, 701–706. [Google Scholar]
- Chatzivassiliou EK and Nolasco G, 2014. Detection of a new variant of Citrus tristeza virus in Greek citrus crops. Phytopathologia Mediterranea, 53, 140–147. [Google Scholar]
- Davino S, Rubio L and Davino M, 2005. Molecular analysis suggests that recent Citrus tristeza virus outbreaks in Italy were originated by at least two independent introductions. European Journal of Plant Pathology, 111, 289–293. [Google Scholar]
- Dawson WO, Garsey SM, Tatineni S, Folimonova SY, Harper SJ and Gowda S, 2013. Citrus tristeza virus–host interactions. Frontiers in Microbiology, 4, 11–20. 10.3389/fmicb.2013.00088. eCollection 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA PLH Panel (EFSA Panel on Plant Health), 2010. PLH Guidance on a harmonised framework for pest risk assessment and the identification and evaluation of pest risk management options by EFSA. EFSA Journal 2010;8(2):1495, 66 pp. 10.2903/j.efsa.2010.1495 [DOI] [Google Scholar]
- EFSA PLH Panel (EFSA Panel on Plant Health), 2014. Scientific Opinion on the pest categorisation of Citrus tristeza virus . EFSA Journal 2014;12(12):3923, 32 pp. 10.2903/j.efsa.2014.3923 [DOI] [Google Scholar]
- EPPO (European and Mediterranean Plant Protection Organization), 2004. Diagnostic protocols for regulated pests, PM 7/31. Citrus tristeza closterovirus. Bulletin OEPP/EPPO Bulletin, 34, 155–157. [Google Scholar]
- EPPO (European and Mediterranean Plant Protection Organization), 2017. EPPO Global Database. Available online: https://gd.eppo.int
- FAO (Food and Agriculture Organization of the United Nations), 2004. ISPM (International Standards for Phytosanitary Measures) 21—Pest Risk Analysis of Regulated Non‐quarantine Pests. FAO, Rome, 30 pp. Available online: https://www.ippc.int/sites/default/files/documents//1323945746_ISPM_21_2004_En_2011-11-29_Refor.pdf [Google Scholar]
- FAO (Food and Agriculture Organization of the United Nations), 2013. ISPM (International Standards for Phytosanitary Measures) 11—Pest Risk Analysis for Quarantine Pests. FAO, Rome, 36 pp. Available online: https://www.ippc.int/sites/default/files/documents/20140512/ispm_11_2013_en_2014-04-30_201405121523-494.65%20KB.pdf [Google Scholar]
- Ferretti L, Fontana A, Sciarroni R, Schimio R, Loconsole G, Albanese G and Saponari M, 2014. Molecular and biological evidence for a severe seedling yellows strain of Citrus tristeza virus spreading in southern Italy. Phytopathologia Mediterranea, 53, 3–13. [Google Scholar]
- Folimonova SY, 2012. Superinfection exclusion is an active virus– controlled function that requires a specific viral protection. Journal of Virology, 86, 5554–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folimonova SY, 2013. Developing an understanding of cross‐protection by Citrus tristeza virus. Frontiers in Microbiology, 4, 76. 10.3389/fmicb.2013.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnsey SM and Cambra M, 1991. Enzyme‐linked immunosorbent assay (ELISA) for citrus pathogens. In: Roistacher CN (ed.). Graft‐transmissible Diseases of Citrus. Handbook for detection and diagnosis. FAO, Rome, Italy, pp. 193–216. [Google Scholar]
- Garnsey SM and Lee RF, 1988. Tristeza. 48–50. In: Whiteside JO, Garnsey SM and Timmer LW (eds.). Compendium of Citrus Diseases. APS Press, St Paul, MN, USA, 80 pp. [Google Scholar]
- Garnsey SM, Permar TA, Cambra M and Henderson CT, 1993. Direct tissue blot immunoassay (DTBIA) for detection of citrus tristeza virus (CTV). In: Moreno P, Da Graça J and Timmer LW (eds.). Proceedings of the 12th Conference of the International Organisation of Citrus Virologists. IOCV, Riverside, CA, USA. pp. 39–50. [Google Scholar]
- Garnsey SM, Civerolo EL, Gumpf DJ, Paul C, Hilf ME, Lee RF, Brlansky R, Yokomi RK and Hartung JS, 2005. Biological characterization of an international collection of Citrus tristeza virus (CTV) Isolates. Proceedings of the 16th Conference of the International Organisation of Citrus Virologists, 2004. IOVC, Riverside, CA, USA, 491, pp. 75–93.
- Gottwald TR, 2010. Concepts in the epidemiology of Citrus tristeza virus. In: Karasev AV and Hilf ME (eds.). Citrus Tristeza Virus Complex and Tristeza Diseases. APS Press, St Paul, MN, USA. pp. 119–131. [Google Scholar]
- Gottwald TR, Cambra M, Moreno P, Camarasa E and Piquer J, 1995. Spatial and temporal analyses of Citrus tristeza virus in Eastern Spain. Phytopathology, 86, 45–55. [Google Scholar]
- Gottwald TR, Garnsey SM, Cambra M, Moreno P, Irey M and Borbón J, 1997. Comparative effects of aphid vector species on increase and spread of citrus tristeza virus. Fruits, 52, 397–404. [Google Scholar]
- Harper SJ, 2013. Citrus tristeza virus: evolution of complex and varied genotypic groups. Frontiers in Microbiology, 4, 2013. 10.3389/fmicb.2013.00093.eCollection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper SJ, Dawson TE and Pearson MN, 2010. Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Archives of Virology, 155, 471–480. [DOI] [PubMed] [Google Scholar]
- Hermoso de Mendoza A, Ballester‐Olmos JF and Pina‐Lorca JA, 1984. Transmission of citrus tristeza virus by aphids (Homoptera, Aphididae) in Spain. In: Garnsey SM, Timmer LW, Dodds JA (eds.). Proceedings of the 9th Conference of the International Organization of Citrus Virologists. IOCV, Riverside, CA, USA. pp. 23–27. [Google Scholar]
- Hilf ME and Garnsey SM, 2002. Citrus tristeza virus in Florida: A synthesis of historical and contemporary biological, serological and genetic data. Proceedings of the 15th Conference of the International Organization of Citrus Virologists. IOCV, Riverside, CA, USA, 13–16.
- de Jong Y, Verbeek M, Michelsen V, Bjørn P, Los W, Steeman F, Bailly N, Basire C, Chylarecki P, Stloukal E, Hagedorn G, Wetzel FT, Glöckler F, Kroupa A, Korb G, Hoffmann A, Häuser C, Kohlbecker A, Müller A, Güntsch A, Stoev P and Penev L, 2014. Fauna Europaea ‐ all European animal species on the web. Biodiversity Data Journal, 2, e4034. 10.3897/BDJ.2.e4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasev VA and Bar‐Joseph M, 2010. Citrus tristeza virus and the taxonomy of Closteroviridae. In: Karasev AV and Hilf ME (eds.). Citrus Tristeza Virus Complex and Tristeza Diseases. APS Press, St Paul, MN, USA. pp. 119–131. [Google Scholar]
- Kitajima EW, Müller GW and Costa AS, 1974. Electron microscopy of tristeza‐infected Passiflora gracilis Jacq. In: Weathers LG and Cohen M (eds.). Proceedings of the 6th Conference of the International Organization of Citrus Virologists. University of California, Division of Agricultural Sciences, Berkeley, USA. pp. 79–82. [Google Scholar]
- Licciardello G, Scuderi G, Ferraro R, Giampetruzzi A, Russo M, Lombardo A, Raspagliesi D, Bar‐Joseph M and Catara A, 2015. Deep sequencing and analysis of small‐RNAs in sweet orange grafted on sour orange infected with two Citrus tristeza virus isolates prevalent in Sicily. Archives of Virology, 160, 2583–2589. [DOI] [PubMed] [Google Scholar]
- Malandraki I, Marouli E and Varveri C, 2011. New isolates of Citrus tristeza virus naturally occurring in old lemon and mandarin trees in Greece. New Disease Reports, 23, 2. [Google Scholar]
- Martelli GP, Agranovsky AA, Bar‐Joseph M, Boscia D, Candresse T, Coutts RHA, Dolja VV, Hu JS, Jelkmann W, Karasev AV, Martin RR, Minafra A, Namba S and Vetten HJ, 2012. Closteroviridae. In: King AMQ, Adams MJ, Carstens EB and Lefkowitz EJ (eds.). Virus Taxonomy, 9th Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, London, UK. pp. 987–1001. [Google Scholar]
- McClean APD, 1957. Tristeza virus of citrus: evidence for absence of seed transmission. The Plant Disease Reporter, 41, 821. [Google Scholar]
- Melzer MJ, Borth WB, Sether DM, Ferreira S, Gonsalves D and Hu JS, 2010. Genetic diversity and evidence for recent modular recombination in Hawaiian Citrus tristeza virus. Virus Genes, 40, 111–118. 10.1007/s11262-009-0409-3. [DOI] [PubMed] [Google Scholar]
- Michaud JP, 1998. A review of the literature on Toxoptera citricida (Kirkaldi) (Homoptera: Aphididae). Florida Entomologist, 81, 37–61. [Google Scholar]
- Moreno P and Garnsey SM, 2010. Citrus Tristeza diseases—A worldwide perspective. In: Karasev AV and Hilf ME (eds.). Citrus Tristeza Virus Complex and Tristeza Diseases. APS Press, St Paul, MN, USA. pp. 27–49. [Google Scholar]
- Moreno P, Ambrós S, Albiach‐Martí MR, Guerri J and Peña L, 2008. Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Molecular Plant Pathology, 9, 251–268. 10.1111/J.1364-3703.2007.00455.X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller GW, Costa AS, Kitajima EW and Camargo IJB, 1974. Additional evidence that tristeza virus multiplies in Passiflora spp. In: Weathers LG and Choen M (eds.). Proceedings of the 6th Conference of the International Organization of Citrus Virologists. University of California, Division of Agricultural Sciences, Berkeley, CA, USA. pp. 75–78. [Google Scholar]
- NCBI (National Center for Biotechnology Information) GenBank database, 2017. Available at: https://www.ncbi.nlm.nih.gov/
- Nolasco G, de Blas C, Torres V and Ponz F, 1993. A method combining immunocapture and PCR amplification in a microtiter plate for the routine diagnosis of plant viruses and subviral pathogens. Journal of Virological Methods, 45, 201–218. [DOI] [PubMed] [Google Scholar]
- Olmos A, Dasí MA, Candresse T and Cambra M, 1996. Print capture PCR: a simple and highly sensitive method for the detection of plum pox virus (PPV) in plant tissues. Nucleic Acids Research, 24, 2192–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen C, Mathioudakis MAG, Gal P, Nol N, Kalliampakou K, Figas A, Bellan A, Iparaguirre A, Rubio L and Livieratos J, 2014. Evolution and molecular epidemiology of Citrus tristeza virus on Crete: recent introduction of a severe strain. Journal of Phytopathology, 162, 1–5. [Google Scholar]
- Papayiannis LC, Santos C, Kyriakou A, Kapari T and Nolasco G, 2007. Molecular characterization of Citrus tristeza virus isolates from Cyprus on the basis of the coat protein gene. Journal of Plant Pathology, 89, 291–295. [Google Scholar]
- Permar TA, Garnsey SM, Gumpf DJ and Lee RF, 1989. A monoclonal antibody that discriminates strains of citrus tristeza virus. Phytopathology, 80, 224–228. [Google Scholar]
- Pina JA, Moreno P, Juarez J, Guerri J, Cambra M, Gorris MT and Navarro L, 2005. A new procedure to index Citrus tristeza virus‐induced decline on sour orange rootstock. Proceedings of the 16th Conference of the International Organisation of Citrus Virologists, 2004. IOCV, Riverside, CA, USA, 491. Plant Disease Reporter, 48, 102–102.
- Raccah B, Loebenstein G and Bar‐Joseph M, 1976. Transmission of citrus tristeza virus by melon aphid. Phytopathology, 66, 1102–1104. [Google Scholar]
- Rizza S, Lombardo A, Nobile G and Catara A, 2007. Biological and molecular characterization of two additional Citrus tristeza virus isolates associated with sour orange inverse pitting rootstock. Journal of Plant Pathology, 89, 57. [Google Scholar]
- Roistacher CN and Bar‐Joseph M, 1987. Transmission of Citrus tristeza virus by Aphis gossypii and by graft inoculationto and from Passiflora spp. Phytophylactica, 19, 179–182. [Google Scholar]
- Roy A and Brlansky RH, 2009. Population dynamics of a Florida Citrus tristeza virus isolate and aphid‐transmitted subisolates: identification of three genotypic groups and recombinants after aphid transmission. Phytopathology, 99, 1297–1306. [DOI] [PubMed] [Google Scholar]
- Roy A, Choudhary N, Hartung JS and Brlansky RH, 2013. The prevalence of the Citrus tristeza virus trifoliate resistance breaking genotype among Puerto Rican isolates. Plant Diseases, 97, 1227–1234. [DOI] [PubMed] [Google Scholar]
- Rubio L, Ayllón MA, Kong P, Fernández A, Polek ML, Guerri J, Moreno P and Falk BW, 2001. Genetic variation of Citrus tristeza virus isolates from California and Spain: evidence for mixed infections and recombination. Journal of Virology, 75, 8054–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz‐Ruiz S, Moreno P, Guerri J and Ambros S, 2006. The complete nucleotide sequence of a severe stem pitting isolate of Citrus tristeza virus from Spain: comparison with isolates from different origins. Archives of Virology, 151, 387–398. [DOI] [PubMed] [Google Scholar]
- Ruiz‐Ruiz S, Moreno P, Guerri J and Ambrós S, 2009. Discrimination between mild and severe Citrus tristeza virus isolates with a rapid and highly specific real‐time reverse transcription‐polymerase chain reaction method using TaqMan LNA probes. Phytopathology, 99, pp. 307–315. [DOI] [PubMed] [Google Scholar]
- Russo M, Scuderi G, Ferraro R, Raspagliesi D, Bar‐Joseph M, Catara A and Licciardello G, 2015. Phenotype and genotype characterization of an unusual Citrus tristeza virus isolate from Sicily. Journal of Plant Pathology, 97(supplement), 34. [Google Scholar]
- Scuderi G, Lombardo A, Raspagliesi D, Russo M, Catara A and Licciardello G, 2016. Development and evaluation of a novel probe microarray for genotyping Citrus tristeza virus using an integrated lab‐on‐chip device. Journal of Plant Pathology, 98, 25–34. [Google Scholar]
- Varveri C, Olmos A, Pina JA and Marroquin CMC, 2014. Biological and molecular characterisation of a distinct Citrus tristeza virus isolate originated from a lemon tree in Greece. Plant Pathology, 64, 792–798. 10.1111/ppa.12308 [DOI] [Google Scholar]
- Vidal E, Yokomi RK, Moreno A, Bertolini E and Cambra M, 2012. Calculation of diagnostic parameters of advanced serological and molecular tissue‐print methods for detection of Citrus tristeza virus. A model for other plant pathogens. Phytopathology, 102, 114–121. 10.1094/PHYTO-05-11-0139 [DOI] [PubMed] [Google Scholar]
- Vives MC, Rubio L, López C, Navas‐Castillo J, Albiach‐Martí MR, Dawson WO, Guerri J, Flores R and Moreno P, 1999. The complete genome sequence of the major component of a mild Citrus tristeza virus isolate. Journal of General Virology, 80, 811–816. [DOI] [PubMed] [Google Scholar]
- Vives MC, Rubio L, Sambade A, Mirkov TE, Moreno P and Guerri J, 2005. Evidence of multiple recombination events between two RNA sequence variants within a Citrus tristeza virus isolate. Virology, 331, 232–237. 10.1016/j.virol.2004.10.037 [DOI] [PubMed] [Google Scholar]
- Wallace JM and Drake RJ, 1951. Recent developments in studies of quick decline and related diseases. Phytopathology, 41, 785–793. [Google Scholar]
- Wang JB, Bozan O, Kwon SJ, Dang T, Rucker T, Yokomi RK, Lee RF, Folimonova SY, Krueger RR, Bash J, Greer G, Diaz J, Serna R and Vidalakis G, 2013. Past and future of a century old Citrus tristeza virus collection: a California citrus germplasm tale. Frontiers in Microbiology, 4, 366. 10.3389/fmicb.2013.00366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomi RK and Garnsey SM, 1987. Transmission of citrus tristeza virus by Aphis gossypii and Aphis citricola in Florida. Phytophylactica, 19, 169–172. [Google Scholar]
- Yokomi RK, Garnsey SM, Civerolo EL and Gumpf D, 1989. Transmission of exotic citrus tristeza isolates by a Florida colony of Aphis gossypii. Plant Disease, 73, 552–556. [Google Scholar]
- Yokomi RK, Lastra R, Stoetzel MB, Damsteegt VD, Lee RF, Garnsey SM, Gottwald TR, Rocha‐Peña MA and Niblett CL, 1994. Establishment of the brown citrus ahid (Homoptera: Aphidide) in Central America and the Caribbean Basin. Journal of Economic Entomology, 88, 1078–1085. [Google Scholar]


