Abstract
EFSA was requested by the European Commission to produce a report on the susceptibility of olive varieties to the Apulian strain of Xylella fastidiosa (subsp. pauca strain CoDiRO, ST53). A systematic literature search identified 21 references providing results of primary research studies on olive plants infected (naturally or artificially) by ST53. From experimental infectivity studies and from surveys in olive orchards, converging lines of evidence indicate tolerance of the Leccino variety to ST53 infections, although no long‐term observations on yield are available yet. While the variety Leccino can become infected with the pathogen, it develops milder symptoms compared to those observed on susceptible varieties (e.g. Cellina di Nardò, Ogliarola salentina). Also, the size of the X. fastidiosa bacterial populations measured in Leccino‐infected plants is lower compared to susceptible olive varieties. Preliminary results show that tolerance or resistance traits can also be found in other olive varieties. New research is now in place in the EU to study the level of susceptibility of many olive varieties to ST53 infections, therefore more relevant results will become available in the coming years.
Keywords: Apulian strain, CoDiRO, cultivar, DistillerSR, olive, resistance, tolerance
1. Introduction
1.1. Background and Terms of Reference as provided by the requestor1
Pursuant to Articles 29 and 31 of Regulation (EC) No 178/20022, European Commission DG SANTE requested the European Food Safety Authority (EFSA) for scientific advice and technical assistance in the field of plant health as regards the regulated harmful organism Xylella fastidiosa (letter dated 30 June 2016 (ref. SANTE/G1/PDR/svi (2016) 3575400)). In particular, pursuant to Article 31 of Regulation (EC) No 178/2002, EFSA was requested to further specify and update the host plants database of X. fastidiosa currently available (EFSA, 2016) taking into account the different X. fastidiosa subspecies and strains (with particular reference to the European isolates), with inclusion of information on non‐susceptible host plants and varieties and negative results of diagnostic tests where available. EFSA was requested to maintain and update this database periodically and to make new releases available on EFSA website, together with a report. The report should specify the list of plants confirmed to be infected by at least two detection methods in field conditions or via vector transmission under experimental conditions and be published at least annually, or according to needs following agreements between our Services. The request is for the period 2016–2020 and the needs for its continuation will be re‐assessed by the end of this period.
With regard to the request to update and maintain the EFSA host plants database of X. fastidiosa, EFSA was requested to deliver by the end of March 2017 a preliminary report on the hosts of the Apulian strain of X. fastidiosa subsp. pauca (letter dated 2 March 2017 (ref. SANTE.G1/DA/as Ares (2017) 1157971)). The report should include information on non‐susceptible host plants and varieties, and negative results of diagnostic tests when available, as specified in the above‐mentioned mandate, and with a particular focus on available scientific information on research results regarding tolerant or resistant olive varieties. EFSA was also requested that the complete new release of the EFSA host plants database of X. fastidiosa, taking into account scientific and technical literature and the up‐to‐date validated information provided by Member States on the other European outbreaks, is delivered by end September 2017.
1.2. Interpretation of the Terms of Reference
Given the time constraints, this report only focuses on the susceptibility of Olea europaea L. varieties3 to the Apulian strain of the pathogen (X. fastidiosa subsp. pauca strain CoDiRO, ST53, hereafter ST53). The review of the other plant species will be included in the new report linked to the update of the EFSA host plant database of Xylella that will be delivered in September 2017.
1.3. Additional information
In order to obtain the most updated overview on the state of the art of current research activities on the topic in the European Union (EU), requests for information were sent via email to:
XF‐ACTORS4 project coordinator (6 March 2017);
POnTE5 project coordinator (6 March 2017);
Department of agriculture, rural development and environment of Apulian Region (8 March 2017);
EUPHRESCO6 network Coordinator (14 March 2017).
Feedback was received in form of a joint report from the two Horizon 2020 research projects (Technical Report by POnTE and XF‐ACTORS, 2017; 14 March 2017) and email replies (from EUPHRESCO on 20 March, from INIAV, Portugal, on 21 March, and from INRA, France, on 23 March).
2. Data and methodologies
A systematic literature search was performed for the extraction of data on susceptibility to ST53 in O. europaea varieties, in line with the systematic literature review approach proposed in the EFSA guidelines (EFSA, 2010). The methodology was adapted to the scope of the mandate and to its implementation with the DistillerSR software (Evidence Partners, Ottawa, Canada). Furthermore, the forms created in DistillerSR for prescreening and data extraction were kept generic (i.e. not only suitable for the data extraction on olive varieties and ST53, but also more general) in order to permit the application of the same methodology and tool to the complete Xylella host plants database, whose update is planned for September 2017.
The process included the following steps (detailed in Appendix A):
Literature search to collect references on the topic (see Section A.1 of Appendix A);
Screening for relevance to exclude references not suitable to the scope (see Section A.2 of Appendix A);
Data extraction from the selected references (see Section A.3 of Appendix A).
All the steps were performed by two reviewers who acted in parallel or in sequence depending on the tasks.
3. Assessment of the susceptibility of Olea europaea L. varieties to Xylella fastidiosa subsp. pauca ST53
3.1. Scope of the assessment
The first outbreak of the quarantine plant pathogen X. fastidiosa in the EU was discovered on olive trees (O. europaea) showing a massive scorching of leaves followed by wilting of entire branches. From its first discovery in a restricted area near the city of Gallipoli in Lecce province (Apulia region, Italy), the disease spread fast, reaching north up to the provinces of Brindisi and Taranto. In October 2013, the first scientific report on the association between the disease, termed olive quick decline syndrome, and the presence of X. fastidiosa was published (Saponari et al., 2013). Since then, the number of plant species found infected (either with or without symptoms) in the Apulian region has been constantly increasing (EFSA, 2015, 2016; EFSA PLH Panel, 2015) together with the expansion of the infected area. While more host plants of X. fastidiosa are continuously reported, the most striking impact of this pathogen is the olive quick decline syndrome. One of the first studies on the biology of X. fastidiosa strain CoDiRO7 and its causal association to the olive quick decline syndrome was funded by EFSA. This pilot project included major Mediterranean crops and some olive varieties and was finalised in March 2016 (Saponari et al., 2016). The experiments and observations initiated during the project are continuing and are now merged in the recent EU‐funded projects (see Section 3.4).
As O. europaea is the most affected crop of the X. fastidiosa outbreak in Apulia, a large part of all research activities over the last years focused on epidemiological studies of X. fastidiosa in olives and on the vectoring capacity of xylem sap‐feeding insects commonly present in the area where infected olive plants are found.
Given the urgency to provide an updated scientific overview on the level of susceptibility of olive varieties to ST53, EFSA conducted a review of literature on this topic.
3.2. Collected literature
The literature search conducted at the end of January 2017 produced 95 references complemented later by four more scientific articles and reports (Section A.1).
During the screening for relevance (Section A.2), 46 articles not focusing on Xylella and/or not describing primary studies were excluded by title/abstract screening (Section A.2.1) while a further 11 reports were removed after full‐text screening because they were not describing in vivo studies (Section A.2.2).
After data extraction (Section A.3), more references were excluded in order to achieve high precision in the final data set for this report. In this phase, eight references were excluded because they did not contain any entry on O. europaea, nine references were excluded because they were not on ST53 and four references were excluded because they only described surveys with negative results.
At the end of the selection process (22 March 2017), 21 papers and reports were retained (full list in Appendix B) for a total number of 128 data extraction forms. The first scientific paper on X. fastidiosa in Italy was published in 2013 (Saponari et al., 2013) and the most recent article was published in an Italian technical magazine on 23 March 2017 (Boscia et al., 2017). A flow diagram on data extraction is shown in Figure 1 (above).
Figure 1.
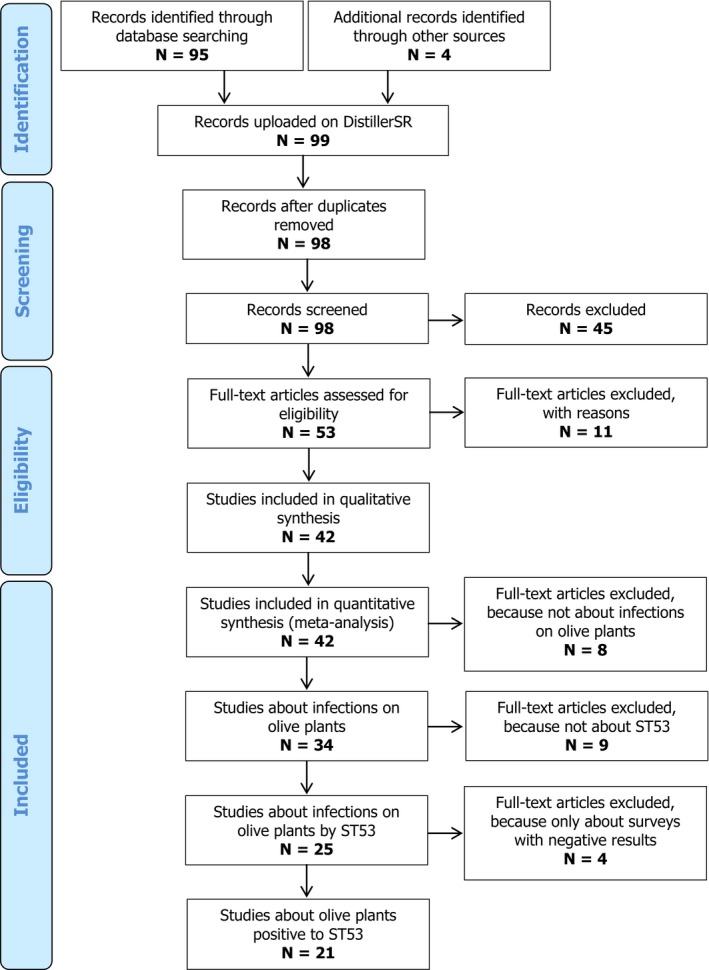
Flow diagram on the data extraction conducted on the DistillerSR tool
3.3. Results
Most references (15) provide results on detection of ST53 in open fields and orchards (study category ‘field survey’). Three references are exclusively dedicated to experimental trials under controlled conditions (study category ‘experimental’) and three cover both categories.
Six papers describe pathogenicity tests and vector transmission studies (natural inoculation) and two reports focus on mechanical inoculation experiments, both conducted by the CNR (Consiglio Nazionale delle Ricerche) of Bari (Saponari et al., 2016; Technical Report by POnTE and XF‐ACTORS, 2017).
Currently, no results are available from long‐term studies over an extended geographical range comprising multiple seasons, due to the fairly recent occurrence and the complex nature of the problem: such studies on tree pathogens are in fact difficult to execute and require long observation times for meaningful conclusions in particular on the host status.
The selected 21 references provided a list of 28 olive varieties in which ST53 was either detected during surveys or developed infections under experimental conditions. Most studies only focus on a few varieties (mainly on Leccino and Ogliarola salentina, followed by Cellina di Nardò, Coratina, Frantoio, Cima di Melfi, Nocellara and Picholine) representing some of the main olive varieties grown in the infected area (Table 1). While the number of reported susceptible varieties is expected to increase with intensified research (Section 3.4), the few studies available also indicate a differential susceptibility of the olive varieties to ST53 (Tables 2 and 3).
Table 1.
Susceptibility of olive varieties to ST53. Host susceptibility information is based on experimental and field survey data
| Variety | Study category | Number of data extraction forms including the variety | Reference number(s) (Appendix B) |
|---|---|---|---|
| Arbequina | Experimental | 3 | [19], [20] |
| Arbosana | Experimental | 3 | [19], [20] |
| Ascolana tenera | Field survey | 1 | [20] |
| Carolea | Field survey | 3 | [2], [20] |
| Cellina di Nardò | Experimental and field survey | 7 | [19], [20] |
| Cima di Melfi | Experimental | 4 | [19], [20] |
| Cipressino | Field survey | 1 | [20] |
| Coratina | Experimental | 7 | [4], [5], [19], [20] |
| Dolce di Cassano | Field survey | 1 | [20] |
| Frantoio | Experimental | 5 | [19], [20] |
| FS‐17® | Field survey | 2 | [2], [20] |
| Gioconda | Field survey | 3 | [2], [20] |
| Kalamata | Field survey | 2 | [2], [20] |
| Koroneiki | Experimental | 3 | [19], [20] |
| Leccino | Experimental and field survey | 15 | [2], [10], [15], [19], [20] |
| Maiatica | Field survey | 1 | [20] |
| Nocellara | Field survey | 4 | [2], [19], [20] |
| Nocellara etnea | Field survey | 1 | [20] |
| Nocellara messinese | Field survey | 1 | [20] |
| Nociara | Field survey | 1 | [20] |
| Ogliarola barese | Experimental | 1 | [20] |
| Ogliarola salentina | Experimental and field survey | 10 | [1], [2], [4], [10], [15], [19], [20] |
| Oliastro | Field survey | 1 | [20] |
| Pendolino | Field survey | 1 | [20] |
| Peranzana | Field survey | 1 | [20] |
| Picholine | Field survey | 4 | [2], [19], [20] |
| Termite di Bitetto | Field survey | 1 | [20] |
| Toscanina | Field survey | 1 | [20] |
| Seedlings (generic) | Experimental | 2 | [19] |
| Not specified | Experimental and field survey | 44 | |
| Total | 134 a |
The total number of forms (see glossary for definition) provided in this table (134) is higher than the one mentioned in Section 3.2 (128) because in two cases a single form included four olive varieties whose figures were represented in the reference as aggregated.
Table 2.
Summary table of ELISA tests (average absorbance OD405nm of positive samples) presented by Boscia et al. (2017) comparing olive varieties' response to ST53 infections observed in different orchards in the infected area
| Location of the survey | FS‐17® | Leccino | Ogliarola salentina |
|---|---|---|---|
| Racale (Lecce province) | 0.875 OD405nm | 1.677 OD405nm | |
| Sannicola (Lecce province) | 0.46 OD405nm | 0.67 OD405nm | 1.69 OD405nm |
| Ugento (Lecce province) | 0.24 OD405nm | 1.40 OD405nm |
ELISA: enzyme‐linked immunosorbent assay.
Table 3.
Summary table of qPCR average values provided by different studies comparing olive varieties' response to ST53 infections
| Trial and reference | Cellina di Nardò | Coratina | Frantoio | FS‐17® | Leccino | Ogliarola salentina |
|---|---|---|---|---|---|---|
| Survey in Sannicola (Lecce province), Boscia et al. (2017) | 5.04 × 104 CFU/mL | 9.93 × 104 CFU/mL | 4.51 × 106 CFU/mL | |||
| Quantification of ST53 population size, Giampetruzzi et al. (2016) | 3.89 × 104 CFU/mL | 2.33 × 106 CFU/mL | ||||
| Artificial inoculation in greenhouse, results after 12 months, Saponari et al. (2016)a |
Stem: 6.77 × 106 CFU/mL Leaf petioles: 3.34 × 106 CFU/mL |
Stem: 5.65 × 105 CFU/mL Leaf petioles: 7.59 × 104 CFU/mL |
Stem: 3.01 × 105 CFU/mL Leaf petioles: 5.18 × 105 CFU/mL |
Stem: 6.38 × 104 CFU/mL Leaf petioles: 3.52 × 105 CFU/mL |
||
| Experimental plot, Technical Report by POnTE and XF‐ACTORS (2017)a | 8.68 × 105 CFU/mL | 7.41 × 105 CFU/mL | 4.95 × 105 CFU/mL | 4.19 × 103 CFU/mL |
qPCR: quantitative polymerase chain reaction; CFU: colony‐forming unit.
Data in the last two rows were kindly provided by the authors of the reference as personal communication.
Most evidence has been collected by research groups located in Apulia.8 From their work, the variety Leccino was identified as tolerant to X. fastidiosa infections (Saponari et al., 2016). From experimental infectivity studies and from surveys in olive orchards, converging lines of evidence indicate tolerance of the Leccino variety to ST53 infections, although long‐term production data are not available yet. While the variety Leccino can become infected with the pathogen, it develops milder symptoms compared to those observed on susceptible varieties (e.g. Cellina di Nardò, Ogliarola salentina) (Giampetruzzi et al., 2016; Boscia et al., 2017). Also, the size of the ST53 bacterial populations measured in infected plants is shown to be two orders of magnitude lower in Leccino than in the susceptible varieties Cellina di Nardò (Saponari et al., 2016; Technical Report by POnTE and XF‐ACTORS, 2017) and Ogliarola salentina (Giampetruzzi et al., 2016; Boscia et al., 2017) (Tables 2 and 3).
Giampetruzzi et al. (2016) also showed that the transcriptome profiles of the two varieties Leccino and Ogliarola salentina upon infection by ST53 were distinctively different, whereas those of the healthy plants were similar. Such findings are consistent with previous studies performed on Vitis vinifera (Choi et al., 2013) or on Citrus sp. (Rodrigues et al., 2013). Further investigations are needed to understand the molecular mechanism underlying such varietal differential response of olive to ST53.
Recent observations on olive trees under field conditions identified the olive variety FS‐17® as an olive genotype with possible resistance traits: in a heavily‐infected multivarietal olive orchard, the average size of ST53 bacterial populations in infected FS‐17® was only about half the size of ST53 bacterial populations observed in infected Leccino plants grown in the same plot (Boscia et al., 2017) (Table 3).
Xylella fastidiosa acquisition efficiency by its vectors is thought to be correlated with the bacterial population size within the plants (Hill and Purcell, 1997). Although the lower ST53 bacterial population size observed in tolerant olive varieties could possibly reduce the vector acquisition efficiency, the occurrence of the disease and the detection of the pathogen in infected plants of these varieties show that vector transmission still occurs. Therefore, it is important that the growth of tolerant olive varieties is accompanied by the implementation of good agriculture practices and integrated pest management for vectors control.
The responses of olive varieties to X. fastidiosa infections are currently further investigated in olive inoculation trials performed in Italy and France by the POnTE and XF‐ACTORS projects (Table 4; Section 3.4). In the experimental design of these trials, the varieties Leccino and Cellina di Nardò are included as tolerant and susceptible controls (Technical Report by POnTE and XF‐ACTORS, 2017).
Table 4.
Ongoing and future inoculation trials with Xylella fastidiosa strains and different host plants as part of POnTE and/or XF‐ACTORS work plans (aggregated information: not all the possible combinations of strain and host species will be tested)
| Country | Institution | Consortium | Xylella strain | Host species |
|---|---|---|---|---|
| Belgium | ILVO | XF‐ACTORS |
|
Nerium oleander, Prunus domestica subsp., Quercus petraea, Salix alba, Vitis vinifera |
| France | ANSES | POnTE |
|
Catharanthus roseus, Citrus sinensis, Coffea arabica, Coffea canephora, Medicago sativa, Nicotiana tabacum |
| France | INRA | POnTE and XF‐ACTORS |
|
Citrus clementina, Citrus maxima, Coffea arabica, Nerium oleander, Olea europaea, Polygala myrtifolia, Vitis vinifera |
| Italy | IPSP‐CNR DiSSPA‐UNIBA | POnTE and XF‐ACTORS |
|
Olea europaea, Vitis vinifera |
| Netherlands | NVWA | XF‐ACTORS |
|
Catharanthus roseus, Coffea sp., Nerium oleander, Nicotiana tabacum, Polygala myrtifolia, Prunus avium, Prunus domestica |
3.4. Ongoing research activity
Given the gravity of the disease caused by X. fastidiosa in olives and the threat it also presents to other crops and other regions in the EU, this pathogen has quickly become a priority research topic in many EU and Mediterranean countries. After the first EFSA funded pilot project (Saponari et al., 2016), two H2020 EU‐funded projects started in the last 2 years: POnTE and XF‐ACTORS. The work plans of both projects allocate major research activities to pathogenicity tests and host range studies for the EU strains of X. fastidiosa, with particular focus on ST53.
Based on the pioneering work on X. fastidiosa conducted by Apulian researchers, more research groups from Belgium, France and the Netherlands are applying similar methodologies (e.g. needle inoculation, vectors transmission) to study the impact of ST53 and other X. fastidiosa strains on various host plant species (Table 4).
In France, INRA is currently conducting inoculation trials on olive varieties Aglanglau, Cailletier, Picholine du Gard and Sabine. A first trial was conducted between 2015 and 2016, involving X. fastidiosa subsp. pauca, CFBP 8402 –ST53 and the variety Cailletier (which is widely cultivated in Corse) but results are not yet available (Technical Report by POnTE and XF‐ACTORS, 2017; personal communication from Marie‐Agnes Jacques, INRA, 23 March 2017).
In Italy, research conducted by IPSP‐CNR DiSSPA‐UNIBA is ongoing to investigate the susceptibility of olive varieties to ST53, based on inoculation trials and ‘sentinel trials’ on four new experimental plantations established in the infected area in southern Apulia. The following (more than 60) olive varieties/selections are included in at least one of these experiments: Arbequina o.p., Arbosana, Ascolana tenera, Ayvalik, Barnea, Bella di Spagna, Biancolilla, Canino/caninese, Carolea, Cassanese, Cellina di Nardò, Changlot Real, Changlot × Dolce Agogia, Chemlal de Kabylie, Chetoui, Cipressino, Coratina, Empeltre, Frantoio o.p., Frantoio × Arbosana, Galega, Giarraffa, Gordal sevillana, Hojiblanca, Itrana, JA5 (O. europaea subsp. europaea var. sylvestris), Kalamon, Koroneiki o.p., Koroneiki × Empeltre, Lastovka, Leccino, Manzanilla de Sevilla, Mastoidis, Megaritiki, Memeçik, Meski, Moraiolo, Morrut, Nocellara del Belice, Nocellara etnea, Nocellara messinese, Nociara, Nolca, Ogliarola barese, Ogliarola salentina, O. europaea subsp. cerasiformis, Olivastro, Ottobratico, Pasola, Picholine, Picholine marrocaine, Picula, Picudo, PicualxJeac9, MSAC43 (O. europaea subsp. europaea var. sylvestris), OLM28 (O. europaea subsp. europaea var. sylvestris), OLM40 (O. europaea subsp. europaea var. sylvestris), Pendolino, Roggianella, San Agostino, Simone, Termite di Bitetto, TN2 (O. europaea subsp. guanchica), Urano, TN10 (O. europaea subsp. guanchica), Toffahi, Uovo di Piccione, Uslu.
In addition more than 200 olive varieties have been grafted in 2016 on infected olive trees to observe the grafts responses to the ST53 infection (Boscia et al., 2017).
Conclusions
EFSA was requested by the European Commission to produce a report on the susceptibility of olive varieties to the Apulian strain of X. fastidiosa (subsp. pauca strain CoDiRO, ST53). A systematic literature search identified 21 references providing results of primary research studies on olive plants susceptibility to ST53.
The main conclusions that can be drawn at this phase are that:
Given the recent introduction of X. fastidiosa into the EU, up to now only limited information on olive varieties tolerant or resistant to X. fastidiosa subsp. pauca ST53 is available.
For the same reason, there are no studies yet to assess the impact of X. fastidiosa subsp. pauca ST53 on yield and quality of olive fruits of the tolerant olive varieties.
From experimental infectivity studies and from surveys in olive orchards, converging lines of evidence indicate tolerance of the olive variety Leccino to X. fastidiosa subsp. pauca ST53 infections. While the variety Leccino can become infected with the pathogen, it develops milder symptoms compared to those observed on susceptible varieties (e.g. Cellina di Nardò, Ogliarola salentina). Also, the size of the X. fastidiosa subsp. pauca ST53 bacterial populations measured in Leccino infected plants is lower compared to susceptible olive varieties.
Preliminary results show that tolerance or resistance traits can also be found in other olive varieties. In observations conducted in a single heavily‐infected multivarietal olive orchard, the average size of X. fastidiosa subsp. pauca ST53 bacterial populations in infected plants of the olive variety FS‐17® was shown to be only about half the size of bacterial populations in infected Leccino plants grown in the same plot.
The occurrence of the disease and the detection of the pathogen in infected plants of these varieties show that the vector transmission still occurs. It is therefore important that the growth of tolerant olive varieties is accompanied by the implementation of good agriculture practices and integrated pest management for vectors control.
New research from the Horizon 2020 research projects POnTE and XF‐ACTORS and nationally funded projects are now in place to study the level of susceptibility of many olive varieties to X. fastidiosa subsp. pauca ST53 infections. It is expected that additional evidence of available sources of resistance will be produced in the coming years from these enhanced research efforts.
Glossary and Abbreviations
- CFU
Colony‐forming unit
- CoDiRO
Complesso del Disseccamento Rapido dell'Olivo (Italian translation of the olive quick decline syndrome)
- DistillerSR
Systematic review software (Evidence Partners, Ottawa, Canada) used for literature data extraction
- ELISA
Enzyme‐linked immunosorbent assay
- Form
In this statement, the term refers to the data collection form used in DistillerSR. It includes information collected from the bibliographic references and represents a unique combination of plant species (and variety, if present), infection determination, location and Xylella identification.
- MLST
Multilocus sequence typing
- qPCR
Quantitative polymerase chain reaction
- Susceptibility
Susceptibility is the sum total of the qualities which make a plant a fit host for the pathogen (Robinson, 1969).
- Systematic literature review
A systematic literature review is a type of literature review of existing evidence pertinent to a clearly formulated question, which uses pre‐specified and standardised methods to identify and critically appraise relevant research, and to collect, report and analyse data from the studies that are included in the review (EFSA, 2010).
- Resistance
Resistance is a term used to describe a host plant response to infection. This strategy inhibits or limits the infection (Roy and Kirchner, 2000), by preventing the infection, or limiting subsequent pathogen growth and development within the host through avoidance or clearance of infection (Miller et al., 2005).
- Tolerance
Tolerance is a term used to describe a host plant response to infection. In a tolerant plant the infection is not inhibited, but its negative fitness consequences are reduced or offset (Roy and Kirchner, 2000) by reducing the additional mortality due to infection or restoring the reproductive ability (fecundity) of infected individuals (Horns and Hood, 2012).
References
EFSA (European Food Safety Authority), 2010. Application of systematic review methodology to food and feed safety assessments to support decision making. EFSA Journal 2010;8(6):1637, 90 pp. doi:10.2903/j.efsa.2010.1637
Horns F and Hood ME, 2012. The evolution of disease resistance and tolerance in spatially structured populations. Ecology and Evolution, 2, 1705–1711.
Miller MR, White A and Boots M, 2005. The evolution of host resistance: tolerance and control as distinct strategies. Journal of Theoretical Biology, 236, 198–207.
Robinson RA, 1969. Disease resistance terminology. Review of Applied Mycology, 48, 593–606.
Roy BA and Kirchner JW, 2000. Evolutionary dynamics of pathogen resistance and tolerance. Evolution, 54, 51–63.
Appendix A – Systematic literature search methodology
A.1. Literature search
The review question (i.e ‘which olive variety is susceptible/tolerant to Xylella fastidiosa ST53?’) was broken down into key elements using the PECO conceptual model (EFSA, 2010), being P (population of interest) the population of Olea europaea varieties in the Apulia region, E (exposure) the exposure to infection by X. fastidiosa, C (comparator) the control/not‐infected plants, and O (outcomes/condition of interest) the plant infection or susceptibility. No more details were added at this phase in order to avoid excluding informative papers.
During the search process, two main elements were considered: the sources of information (databases) to be consulted (Table A.1) and the development of search strategies (Table A.2).
Table A.1.
Sources of information
| Database | Time coverage | Platform |
|---|---|---|
| Web of Science Core Collection | 1975–present | Web of Science |
| CABI: CAB Abstracts | 1910–present | Web of Science |
| BIOSIS Citation Index | 1926–present | Web of Science |
| Chinese Science Citation Database | 1989–present | Web of Science |
| Current Contents Connect | 1998–present | Web of Science |
| Data Citation Index | 1900–present | Web of Science |
| FSTA | 1969–present | Web of Science |
| KCI‐Korean Journal Database | 1980–present | Web of Science |
| Russian Science Citation Index | 2005–present | Web of Science |
| MEDLINE | 1950–present | Web of Science |
| SciELO Citation Index | 1997–present | Web of Science |
| Zoological Record | 1864–present | Web of Science |
| Scopus | 1960–present | Elsevier |
| Pubmed | 1946–present | NCBI |
Table A.2.
Search strings used in all the selected sources of information
| Search strings | Databases | ||
|---|---|---|---|
| Web of Science | Scopus | PubMed | |
| “Xylella” AND “oliv*” | 85 | 44 | 16 |
| “Xylella” AND “ital*” | 113 | 46 | 22 |
| “Xylella” AND “Apulia” | 40 | 22 | 5 |
| “Xylella” AND “Puglia” | 25 | 2 | 0 |
| “CoDiRO” OR “ST53” | 30 | 14 | 5 |
| “Olive quick decline syndrome” | 30 | 17 | 7 |
The search strategies were designed combining separated searches (splitting) on the two main aspects relevant to the scope: pathogen and host OR pathogen and location (i.e. exposure (E) and population of interest (P)).
The defined search strategies (Table A.2) were run in all the selected information sources (Table A.1) on 27 January 2017. No language, date or document type restrictions were applied in the search strings.
The collected records were downloaded and imported into EndNote® X8 bibliographic management software. Duplicate entries were removed by the same software, leaving 95 final records which were uploaded on DistillerSR together with the full texts in portable document format (pdf).
Four additional references were included at a later stage because i) they were published after the 27 January 2017 (three titles), or ii) they were unpublished reports (one title).
Finally, one reference was excluded by the Duplicate Detection feature of DistillerSR (Figure 1).
A.2. Screening for relevance
The collected references were screened for relevance in two sequential steps:
Title/abstract screening of all the references;
Full‐text screening of those references which were not excluded during the previous step.
Specific inclusion/exclusion criteria were applied at each step (Tables A.3 and A.4, respectively).
Table A.3.
Inclusion criteria for the title/abstract screening
| Question text | Type of answer | Answer text | Exclusion criteria |
|---|---|---|---|
| Is Xylella the topic of the study? | Only one of the possible alternative answers can be selected | Yes | Included |
| No | Excluded | ||
| Is it a primary research study? | Only one of the possible alternative answers can be selected | Yes | Included |
| No | Excluded |
Table A.4.
Inclusion criteria for the full text screening
| Question text | Type of answer | Answer text | Exclusion criteria |
|---|---|---|---|
| Is an English abstract present? | Only one of the possible alternative answers can be selected | Yes | Neutral |
| No | Neutral | ||
| Which is the type of the publication? | Only one of the possible alternative answers can be selected | Peer‐reviewed article | Neutral |
| Article | Neutral | ||
| Book | Neutral | ||
| Conference proceedings | Neutral | ||
| Abstract | Neutral | ||
| Technical papers/Report | Neutral | ||
| Genbank/MLST database/any sequence database | Neutral | ||
| Other | Neutral | ||
| Is Xylella presence investigated in vivo in the plant? | Only one of the possible alternative answers can be selected | Yes | Included |
| No | Excluded |
The two reviewers worked in parallel: whenever the software identified a discrepant reply by the two reviewers on a given reference, the review process was stopped until agreement was reached.
A.2.1. Title/abstract screening
The first step required the reviewers to reply to two questions (Table A.3) having considered the title and abstract (if available) of the reference but not the full text. The aim of this step was to guarantee the inclusion in the data extraction only of papers presenting original research data (i.e. primary research studies) on Xylella. Literature reviews on the topic were therefore excluded.
A negative reply to one of the two questions, if confirmed by the second reviewer, excluded the reference. In case the information provided in title/abstract was insufficiently clear, the reviewers accepted the reference and passed it to the full text screening for further consideration.
A.2.2. Full‐text screening
All papers which passed the title/abstract screening were subjected to the full‐text screening. This step required the reviewers to reply to three questions (Table A.4): the first two having a descriptive scope (neutral) and the third question having an inclusion/exclusion role. The descriptive questions were added at this phase in order to collect information about the type of reference but not to exclude a reference. For example, not only peer‐reviewed journals were accepted, since some of the recent findings on X. fastidiosa could first be presented in national technical magazines or reports. Regarding the third question, only papers describing infections of the pathogen observed in the plant (i.e. in vivo) were retained and brought to the data extraction phase. All studies describing in vitro results were therefore excluded.
A.3. Data extraction
For each reference, one or more data extraction forms were filled in to extract all data reported in the study. Each form represents a unique combination of plant species (and variety, if present), infection determination, location and Xylella identification. A form can include more than one plant variety or more than one Xylella strain only when aggregated data were used (see footnote of Table 1 in the statement).
The two reviewers worked in sequence: one reviewer performed the data extraction and the second conducted assessment and quality control of the filled forms.
A.3.1. Data extraction form
Botanical identification of the plant
Plant family
Plant species
Cultivar/variety
Infection determination
Experimental/Survey
Year
Location
Country of origin of the plant
Location: name of the site/area
Latitude: in WGS84, in decimal format
Longitude: in WGS84, in decimal format
Xylellaidentification
Species
Subspecies
Strain/MLST
Isolate
Methods applied
Description of method/s used.
Possible selection of one or more of the following techniques:
Observation of symptoms
Culture method
Pathogenicity test with mechanical inoculation
Pathogenicity tests with vectors
Optical microscope
Fluorescence microscope
Electron microscopy tests
Immunoblotting (IB)
Immunofluorescence method
Enzyme‐linked immunosorbent assay (ELISA)
Polymerase chain reaction (PCR)
Molecular markers
Real time PCR
Quantitative PCR (qPCR)
Sequence analysis
Southern blot
Northern blot
Western blot
Observation of Xylella movement into the plant
Localized/Systemic (if reported)
Indication about the Xylella resistance status of the plant
Tolerant/Resistant (if reported)
Comments
Appendix B – List of references retained by systematic literature search
1.
Bleve G, Marchi G, Ranaldi F, Gallo A, Cimaglia F, Logrieco AF, Mita G, Ristori J and Surico G, 2016. Molecular characteristics of a strain (Salento‐1) of Xylella fastidiosa isolated in Apulia (Italy) from an olive plant with the quick decline syndrome. Phytopathologia Mediterranea, 55, 139–146.
Boscia D, Altamura G, Di Carolo M, Dongiovanni C, Fumarola G, Giampetruzzi A, Greco P, La Notte P, Loconsole G, Manni F, Melcarne G, Montilon V, Morelli M, Murrone N, Palmisano F, Pollastro P, Potere O, Roseti V, Saldarelli P, Saponari A, Saponari M, Savino V, Silletti MR, Specchia F, Susca L, Tauro D, Tavano D, Venerito P, Zicca S and Martelli GP, 2017. Resistenza a Xylella fastidiosa in olivo: stato dell'arte e prospettive. Informatore Agrario, 11, 59–63.
Cariddi C, Saponari M, Boscia D, De Stradis A, Loconsole G, Nigro F, Porcelli F, Potere O and Martelli GP, 2014. Isolation of a Xylella fastidiosa strain infecting olive and oleander in Apulia, Italy. Journal of Plant Pathology, 96, 425–429.
Cornara D, Cavalieri V, Dongiovanni C, Altamura G, Palmisano F, Bosco D, Porcelli F, Almeida RPP and Saponari M, 2016. Transmission of Xylella fastidiosa by naturally infected Philaenus spumarius (Hemiptera, Aphrophoridae) to different host plants. Journal of Applied Entomology, 141, 80–87.
Cornara D, Saponari M, Zeilinger AR, de Stradis A, Boscia D, Loconsole G, Bosco D, Martelli GP, Almeida RPP and Porcelli F, 2017. Spittlebugs as vectors of Xylella fastidiosa in olive orchards in Italy. Journal of Pest Science, 90, 521–530.
Djelouah K, Frasheri D, Valentini F, D'Onghia AM and Digiaro M, 2014. Direct tissue blot immunoassay for detection of Xylella fastidiosa in olive trees. Phytopathologia Mediterranea, 53, 559−564.
Elbeaino T, Valentini F, Abou Kubaa R, Moubarak P, Yaseen T and Digiaro M, 2014. Multilocus sequence typing of Xylella fastidiosa isolated from olive affected by “olive quick decline syndrome” in Italy. Phytopathologia Mediterranea, 53, 533–542.
Elbeaino T, Yaseen T, Valentini F, Ben Moussa IE, Mazzoni V and D'Onghia AM, 2014. Identification of three potential insect vectors of Xylella fastidiosa in southern Italy. Phytopathologia Mediterranea, 53, 328–332.
Giampetruzzi A, Chiumenti M, Saponari M, Donvito G, Italiano A, Loconsole G, Boscia D, Cariddi C, Martelli GP and Saldarelli P, 2015. Draft genome sequence of the Xylella fastidiosa CoDiRO strain. Genome Announcements, 3, e01538‐14. doi: 10.1128/genomeA.01538‐14.
Giampetruzzi A, Morelli M, Saponari M, Loconsole G, Chiumenti M, Boscia D, Savino V, Martelli GP and Saldarelli P, 2016. Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca. BMC Genomics, 17, 475. doi: 10.1186/s12864‐016‐2833‐9.
Guan W, Shao J, Elbeaino T, Davis RE, Zhao T and Huang Q, 2015. Specific detection and identification of American mulberry‐infecting and Italian olive‐associated strains of Xylella fastidiosa by polymerase chain reaction. PLoS One, 10, e0129330.
Loconsole G, Potere O, Boscia D, Altamura G, Djelouah K, Elbeaino T, Frasheri D, Lorusso D, Palmisano F, Pollastro P, Silletti MR, Trisciuzzi N, Valentini F, Savino V and Saponari M, 2014. Detection of Xylella fastidiosa in olive trees by molecular and serological methods. Journal of Plant Pathology, 96, 7–14.
Loconsole G, Saponari M, Boscia D, D'Attoma G, Morelli M, Martelli GP, Almeida RPP, 2016. Intercepted isolates of Xylella fastidiosa in Europe reveal novel genetic diversity. European Journal of Plant Pathology, 146, 85–94.
Mang SM, Frisullo S, Elshafie HS and Camele I, 2016. Diversity evaluation of Xylella fastidiosa from infected olive trees in Apulia (Southern Italy). The Plant Pathology Journal, 32, 102–111.
National Research Council, Institute for Sustainable Plant Protection, 2016. Olea europaea cultivar: Leccino, Ogliarola salentina Transcriptome or Gene expression. European Nucleotide Archive. Available online: http://www.ebi.ac.uk/ena/data/view/PRJNA316374
National Research Council, Institute for Sustainable Plant Protection, 2014. Xylella fastidiosa strain: CoDiRO Genome sequencing. European Nucleotide Archive. Available online: http://www.ebi.ac.uk/ena/data/view/PRJNA269016
Saponari M, Boscia D, Nigro F and Martelli GP, 2013. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy). Journal of Plant Pathology, 95, 659–668.
Saponari M, Loconsole G, Cornara D, Yokomi RK, De Stradis A, Boscia D, Bosco D, Martelli GP, Krugner R and Porcelli F, 2014. Infectivity and transmission of Xylella fastidiosa by Philaenus spumarius (Hemiptera: Aphrophoridae) in Apulia, Italy. Journal of Economic Entomology, 107, 1316–1319.
Saponari M, Boscia D, Altamura G, D'Attoma G, Cavalieri V, Loconsole G, Zicca S, Dongiovanni C, Palmisano F, Susca L, Morelli M, Potere O, Saponari A, Fumarola G, Di Carolo M, Tavano D, Savino V and Martelli P, 2016. Pilot project on Xylella fastidiosa to reduce risk assessment uncertainties. EFSA supporting publication 2016: EN‐1013. 60 pp.
Technical Report by POnTE and XF‐ACTORS, 2017. Institute for Sustainable Plant Protection, CNR, Bari (Italy) with the contributions of the members of the consortium POnTE (635646) and XF‐ACTORS (727987). Studies on the host plants of Xylella fastidiosa in Europe. Provided to EFSA following official request on the 14 March 2017.
Yaseen T, Drago S, Valentini F, Elbeaino T, Stampone G, Digiaro M and D'Onghia AM, 2015. On‐site detection of Xylella fastidiosa in host plants and in “spy insects” using the real‐time loop‐mediated isothermal amplification method. Phytopathologia Mediterranea, 54, 488–496.
Suggested citation: EFSA (European Food Safety Authority) , Baù A, Delbianco A, Stancanelli G and Tramontini S, 2017. Statement on susceptibility of Olea europaea L. varieties to Xylella fastidiosa subsp. pauca ST53: systematic literature search up to 24 March 2017. EFSA Journal 2017;15(4):4772, 18 pp. doi: 10.2903/j.efsa.2017.4772
Requestor: European Commission
Question number: EFSA‐Q‐2016‐00445
Acknowledgements: EFSA wishes to thank the following for the support provided to this scientific output: Bragard C and Winter S for their kind review, Muñoz Guajardo I (EFSA) for her support on systematic literature searches. EFSA wishes to acknowledge all European competent institutions, Member State bodies and other organisations that provided data for this scientific output.
Approved: 29 March 2017
This output was previously in March 2017 as part of EFSA's crisis communication procedures.
Notes
Submitted by European Commission, ref. SANTE/G1/DA/as Ares (2017) 1157971.
Regulation (EC) No 178/2002 of the European Parliament and of the Council of 28 January 2002 laying down the general principles and requirements of food law, establishing the European Food Safety Authority and laying down procedures in matters of food safety, OJ L 31, 1.2.2002, p. 1–24, as last amended.
In support to the reader, in this report only the term ‘variety’ has been used, consistently with the terms of reference; however, ‘cultivar’ would have been equally acceptable.
Xylella fastidiosa subsp. pauca strain CoDiRO belongs to the single sequence type ST53 and to date is the only sequence type present in the Apulian region (EFSA PLH Panel, 2016).
Dipartimento di Scienze del Suolo, della Pianta e degli Alimenti (DiSSPA), Università degli Studi di Bari (DiSSPA‐UNIBA), Aldo Moro Centro di Ricerca, Sperimentazione e Formazione in Agricoltura – Basile Caramia, Locorotondo, Bari Istituto per la Protezione Sostenibile delle Piante (IPSP), Consiglio Nazionale delle Ricerche (CNR), Unità Organizzativa di Bari.
References
- Boscia D, Altamura G, Di Carolo M, Dongiovanni C, Fumarola G, Giampetruzzi A, Greco P, La Notte P, Loconsole G, Manni F, Melcarne G, Montilon V, Morelli M, Murrone N, Palmisano F, Pollastro P, Potere O, Roseti V, Saldarelli P, Saponari A, Saponari M, Savino V, Silletti MR, Specchia F, Susca L, Tauro D, Tavano D, Venerito P, Zicca S and Martelli GP, 2017. Resistenza a Xylella fastidiosa in olivo: stato dell'arte e prospettive. Informatore Agrario, 11, 59–63. [Google Scholar]
- Choi HK, Iandolino A, Silva FG and Cook DR, 2013. Water deficit modulates the response of Vitis vinifera to the Pierce's disease pathogen Xylella fastidiosa . Molecular Plant‐Microbe Interactions, 26, 643–657. [DOI] [PubMed] [Google Scholar]
- EFSA (European Food Safety Authority), 2010. Application of systematic review methodology to food and feed safety assessments to support decision making. EFSA Journal 2010;8(6):1637, 90 pp. doi: 10.2903/j.efsa.2010.1637 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2015. Categorisation of plants for planting, excluding seeds, according to the risk of introduction of Xylella fastidiosa . EFSA Journal 2015;13(3):4061, 31 pp. doi: 10.2903/j.efsa.2015.4061 [DOI] [Google Scholar]
- EFSA (European Food Safety Authority), 2016. Scientific report on the update of a database of host plants of Xylella fastidiosa: 20 November 2015. EFSA Journal 2016;14(2):4378, 40 pp. doi: 10.2903/j.efsa.2016.4378 [DOI] [Google Scholar]
- EFSA PLH Panel (EFSA Panel on Plant Health), 2015. Scientific opinion on the risks to plant health posed by Xylella fastidiosa in the EU territory, with the identification and evaluation of risk reduction options. EFSA Journal 2015;13(1):3989, 262 pp. doi: 10.2903/j.efsa.2015.3989 [DOI] [Google Scholar]
- EFSA PLH Panel (EFSA Panel on Plant Health), 2016. Statement on diversity of Xylella fastidiosa subsp. pauca in Apulia. EFSA Journal 2016;14(8):4542, 19 pp. doi: 10.2903/j.efsa.2016.4542 [DOI] [Google Scholar]
- Giampetruzzi A, Morelli M, Saponari M, Loconsole G, Chiumenti M, Boscia D, Savino V, Martelli GP and Saldarelli P, 2016. Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca . BMC Genomics, 17, 475. doi: 10.1186/s12864-016-2833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill BL and Purcell AH, 1997. Populations of Xylella fastidiosa in plants required for transmission by an efficient vector. Phytopathology, 87, 1197–1201. [DOI] [PubMed] [Google Scholar]
- Rodrigues CM, de Souza AA, Takita MA, Kishi LT and Machado MA, 2013. RNA‐Seq analysis of Citrus reticulata in the early stages of Xylella fastidiosa infection reveals auxin‐related genes as a defense response. BMC Genomics, 14, 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponari M, Boscia D, Nigro F and Martelli GP, 2013. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy). Journal of Plant Pathology, 95, 659–668. [Google Scholar]
- Saponari M, Boscia D, Altamura G, D'Attoma G, Cavalieri V, Loconsole G, Zicca S, Dongiovanni C, Palmisano F, Susca L, Morelli M, Potere O, Saponari A, Fumarola G, Di Carolo M, Tavano D, Savino V and Martelli P, 2016. Pilot project on Xylella fastidiosa to reduce risk assessment uncertainties. EFSA supporting publication 2016: EN‐1013. 60 pp. doi: 10.2903/sp.efsa.2016.EN-1013 [DOI]
- Technical Report by POnTE and XF‐ACTORS , 2017. Institute for Sustainable Plant Protection, CNR, Bari (Italy) with the contributions of the members of the consortium POnTE (635646) and XF‐ACTORS (727987). Studies on the host plants of Xylella fastidiosa in Europe. Provided to EFSA following official request on the 14 March 2017.


