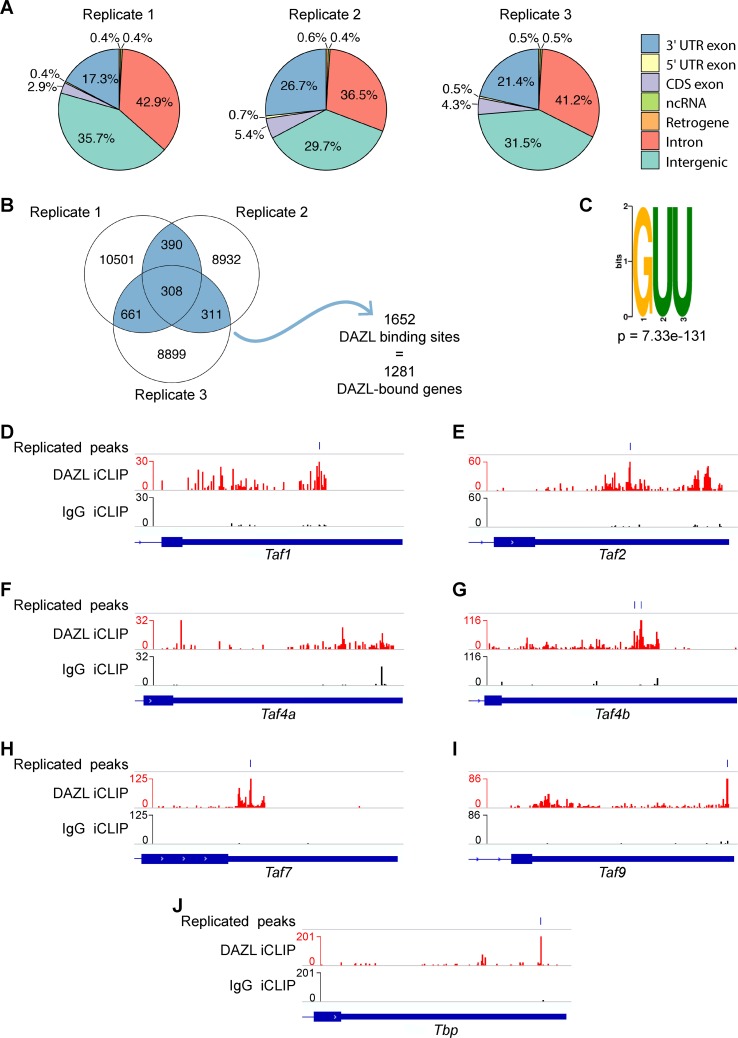
Fig 8. Identification of DAZL targets via iCLIP in testes synchronized for leptotene spermatocytes.
(A) Genomic distribution of DAZL iCLIP peaks identified in 3 biological replicates (p < 0.001). Peaks in each type of genomic region were called via ASPeak. (B) Venn diagram showing overlap of DAZL iCLIP peaks in 3’ UTRs among 3 biological replicates. 1,670 replicated peaks (present in at least 2 of 3 replicates; highlighted in blue) were identified. After merging replicated peaks falling on consecutive nucleotides, 1,652 binding sites were identified. These binding sites correspond to 1,281 genes, which were designated as DAZL-bound genes. (C) GUU motif is enriched at DAZL 3’ UTR peaks. AME from the MEME Suite was used to identify motif enrichment at crosslinked nucleotides in replicated peaks relative to shuffled control sequences. (D-J) DAZL and IgG iCLIP gene tracks showing 3’ UTRs for Taf1 (D), Taf2 (E), Taf4a (F), Taf4b (G), Taf7 (H), Taf9b (I), and Tbp (J). Each iCLIP track represents the crosslinked sites from the sum of unique reads from 3 biological replicates, with DAZL iCLIP reads in red and IgG iCLIP reads in black. The horizontal blue lines on top mark the replicated peaks.