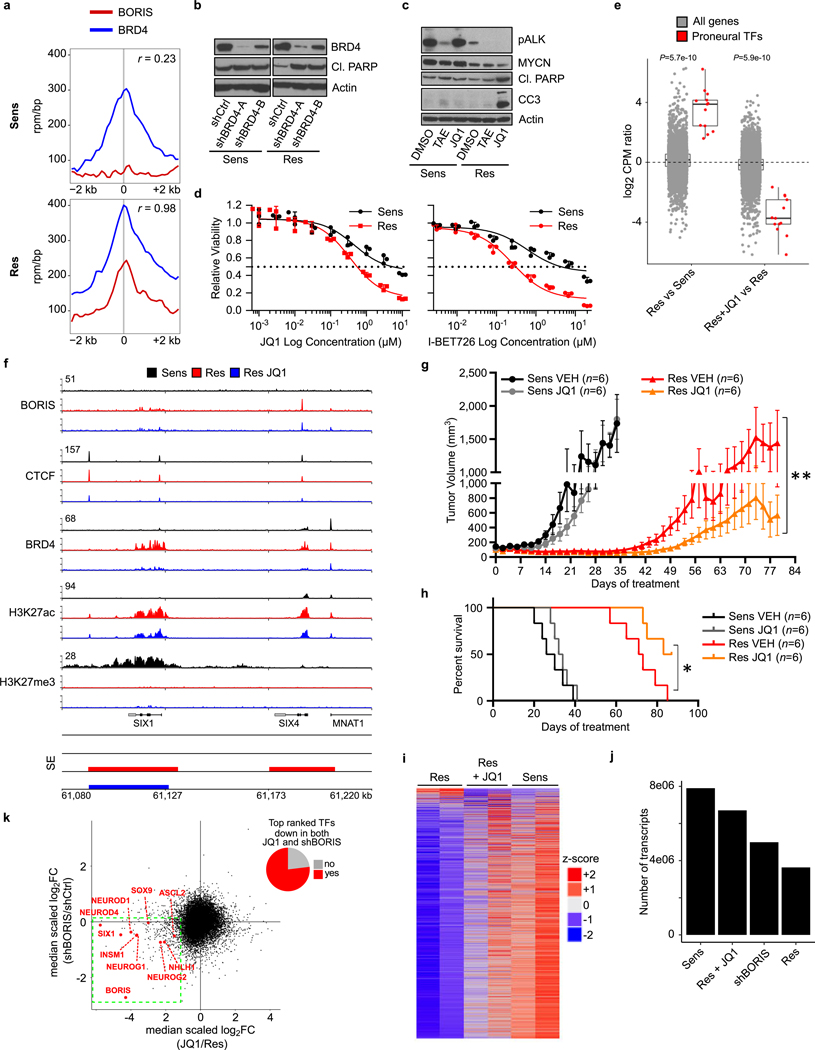
Extended Data Fig. 9 |. The proneural transcription factor network in resistant cells is sensitive to BRD4 inhibition.
a, Metaplots showing the correlation between BRD4 and BORIS co-occupancies at the promoter regions (± 2 kb) of the 89 top-ranked genes in resistant versus sensitive cells based on the features in Fig. 4b (r, Spearman correlation coefficient). b, Immunoblot analysis of BRD4 and cleaved PARP expression in sensitive and resistant cells expressing control (shCtrl) or BRD4 (shBRD4-A and -B) shRNAs. c, Immunoblot analysis of the indicated proteins in sensitive and resistant cells treated with DMSO, TAE684 (1 μM) or JQ1 (2.5 μM) for 48 h. d, Dose–response curves for sensitive and resistant cells treated with JQ1 or I-BET726 (JQ1 (IC50 values: sensitive, 4,798 nM; resistant, 645 nM); I-BET726 (IC50 values: sensitive, 6,203 nM; resistant, 347 nM)). Data are mean ± s.d., n = 3 biological replicates. e, Box plots comparing the expression of the transcription factors listed in Fig. 4b (n = 13) with that of all genes (n = 18,038) in sensitive versus resistant cells (left), and between DMSO and JQ1-treated resistant cells (right) (P values determined by two-sided Wilcoxon rank-sum test). f, ChIP–seq tracks of the indicated proteins at the SIX1 or SIX4 locus in sensitive, resistant and JQ1-treated resistant cells (2.5 μM for 48 h). Super-enhancers are depicted as coloured rectangles below the tracks. Signal intensity is shown in the top left corner for each track. g, h, Tumour volumes (g) and survival curves (h) in sensitive- and resistant-cell xenografts in NU/NU (Crl:NU-Foxn1nu) mice treated with JQ1 (50 mg kg−1 i.p. once daily) and vehicle control for up to 87 days. Data are mean ± s.e.m., n = 6 per arm. Significance was calculated by Mann–Whitney U test for tumour volumes (sensitive: P = 0.3231; resistant: P = 0.0023) and by log-rank test for Kaplan–Meier survival analysis (sensitive: P = 0.3047; resistant: 0.0348), both two-sided. i, Heat map of gene expression values in sensitive, resistant and JQ1-treated resistant cells. Rows are z-scores calculated for each gene in each condition. j, Number of transcripts in sensitive, JQ1-treated resistant, shBORIS-expressing resistant and resistant cells based on expression array data after spike-in normalization. k, Scatter plot displaying the medianscaled fold-change gene expression values for shBORIS and JQ1-treated resistant cells. The top-ranked transcription factors that show decreased expression levels after both BORIS knockdown and JQ1 treatment are listed in red (bottom left quadrant). The pie chart represents the fraction of all top-ranked transcription factors that are located in the left lower quadrant of the scatter plot. All box plots are as defined in Fig. 4. In b, c and f, data are representative of two independent experiments. In a, e and i–k, data are from n = 2 biological replicates (see Supplementary Note 2 for further details; for gel source data, see Supplementary Fig. 1).