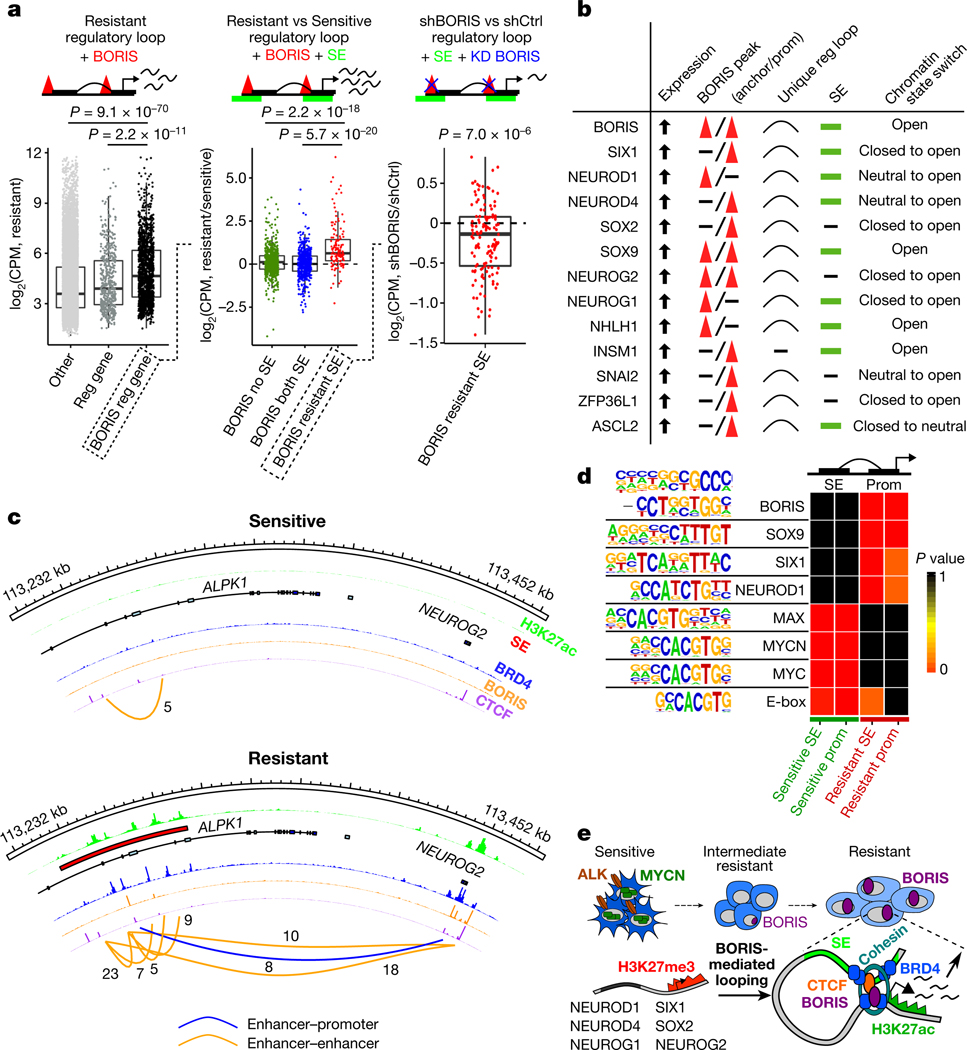
Fig. 4 |. BORIS-regulated chromatin remodelling supports a phenotypic switch that maintains the resistant state.
a, Left, fold change in expression in counts per million (CPM) of genes involved in resistant cell-specific regulatory interactions that are positive for BORIS binding (n = 1,368) versus those involved in regulatory interactions that are negative for BORIS binding (n = 519) or not associated with a new regulatory interaction (other) (n = 16,151). Centre, fold change in expression of genes involved in resistant cell-specific regulatory interactions positive for BORIS binding and associated with super-enhancers (SEs) specific to resistant cells (n = 134) versus those with super-enhancers shared by both cell types (n = 514) or not associated with super-enhancers (n = 720). Right, fold change in expression of genes involved in resistant cell-specific regulatory interactions positive for BORIS binding and associated with resistant cell-specific super-enhancers before and after BORIS knockdown (KD) (n = 134) (P values determined by two-sided Wilcoxon rank-sum test). For all box plots, centre lines denote medians; box limits denote twenty-fifth and seventy-fifth percentiles; whiskers denote minima and maxima (1.5× the interquartile range). b, Highest-ranked transcription factors associated with the resistance phenotype selected based on the presence of at least four of the five indicated features. c, ChIP–seq tracks of the indicated proteins in sensitive and resistant cells at the NEUROG2 locus; regulatory interactions with PET numbers indicated below. d, Transcription factor recognition motifs at super-enhancers and promoters (± 2 kb) of the 1,000 highest-expressed genes in resistant and sensitive cells (n = 2 biological replicates) (P values determined by hypergeometric enrichment test). Panels a–c integrate data of biological replicates from expression microarrays (n = 2), ChIP–seq (n = 2) and HiChIP (n = 3). e, Proposed role of BORIS in resistant cells.