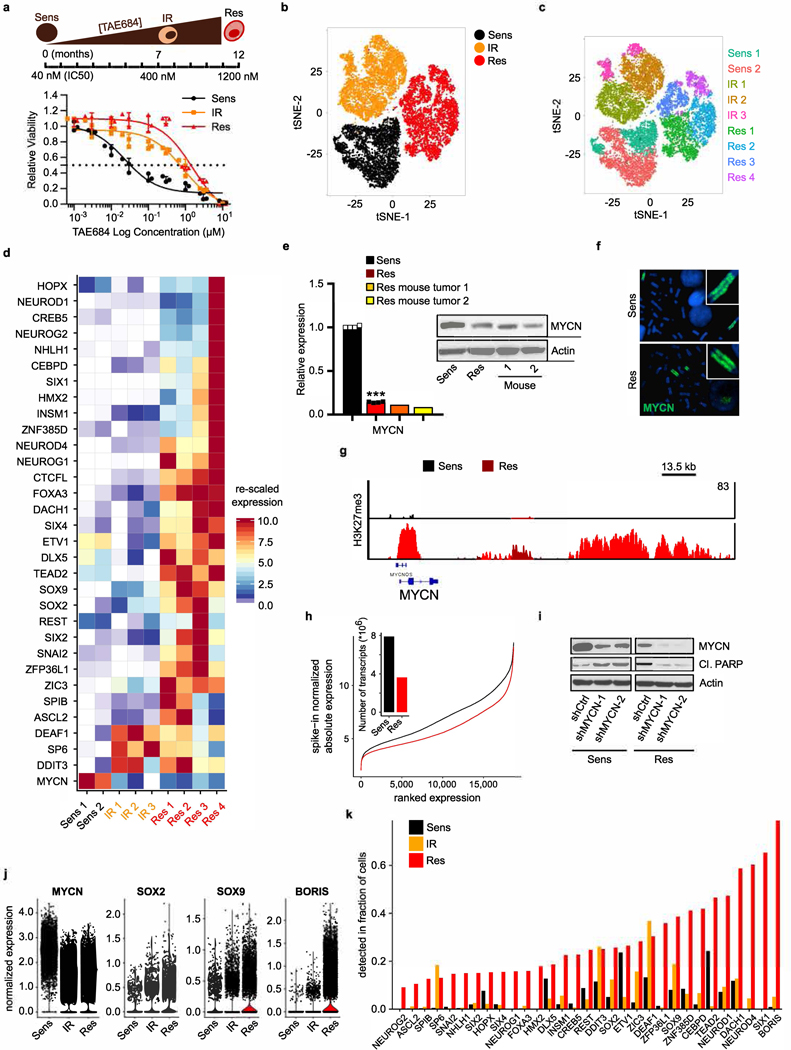
Extended Data Fig. 3 |. Development of resistance is associated with loss of MYCN followed by gradual induction of proneural transcription factors.
a, TAE684 dose–response curves of Kelly neuroblastoma cells during resistance establishment (IC50 values: sensitive, 39.4 nM; intermediate, 618 nM; resistant, 1,739 nM). Data are mean ± s.d., n = 3 biological replicates. Schematic representation of development of resistance is shown above. b, t-SNE plot of scRNA-seq data showing the segregation of sensitive (n = 5,432), intermediate (n = 6,376) and resistant (n = 6,379) cells. c, t-SNE plot depicting unsupervised clusters for the individual subpopulations that underlie the pseudotime analysis. d, Heat map of rescaled gene expression values of the most variable ranked transcription factors in the three cell states. e, qRT–PCR and immunoblot analysis of MYCN expression in TAE684-resistant xenograft tumours (1 and 2) and sensitive and resistant cells in culture. The qRT–PCR data are mean ± s.d., n = 4 biological replicates for sensitive and resistant cells (***P = 1.396 × 10−11; unpaired two-sided t-test) and n = 3 technical replicates for each tumour. f, Fluorescence in situ hybridization of MYCN in sensitive and resistant cells (representative of 20 nuclei per condition). g, ChIP–seq track of H3K27me3 binding at the MYCN locus in sensitive and resistant cells. Signal intensity is given in the top right corner. h, Line plot showing the association between genes ordered by expression (x axis) and changes in absolute gene expression levels (y axis) in sensitive versus resistant cells. Bar plot, total transcriptional yield in sensitive or resistant cells. i, Immunoblot analysis of the indicated proteins in sensitive and resistant cells expressing control (shCtrl) or MYCN (shMYCN-1 and −2) shRNAs. j, Violin plots representing the expression distribution of selected genes in the same cells as in a (centre line, median). k, Bar plot showing the fractions of cells with detectable mRNA levels of the same genes as in d. In e (immunoblot) and f–i, data are representative of two independent experiments (for gel source data, see Supplementary Fig. 1).