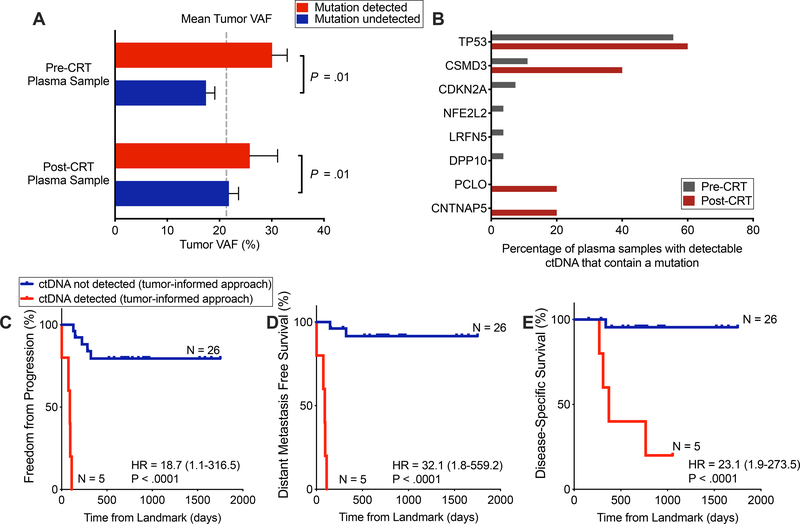
Figure 2. Detection of tumor-informed mutations following chemoradiotherapy is strongly prognostic.
(A) Tumor variant allele fraction (VAF), stratified by whether and at which time point a patient had detectable ctDNA (x-axis categories) and further stratified by whether a given mutation was detectable in plasma (column colors). P value calculated by a 2-way ANOVA and was significant for difference based on if a mutation was detected in plasma (P = .01), but not for if a patient was detected pre- or post-CRT. (B) Tumor mutations detected in plasma pre- and post-CRT. Denominator is the total number of patients with detectable ctDNA, pre- and post-CRT (n=27 and n=5, respectively). Genes depicted were mutated in more than 5% of tumors in cBioportal ESCA datasets. (C-E) Kaplan–Meier analyses comparing patients with detectable and undetectable ctDNA in the post-CRT sample using tumor-informed ctDNA detection for (C) freedom from progression (P<.0001, HR = 18.7 (95%CI, 1.1–316.5)), (D) distant metastasis-free survival (P<.0001, HR = 32.1 (95%CI, 1.8–559.2)) and (E) disease-specific survival (P < .0001, HR = 23.1 (95%CI, 2.0–273.5)). ctDNA negative (n =26) versus ctDNA positive (n = 5). Time (days) was measured from the landmark (post-CRT blood draw). One patient was excluded from this analysis due to an event that occurred prior to the post-CRT blood collection. P values and hazard ratios were calculated from the log-rank test.