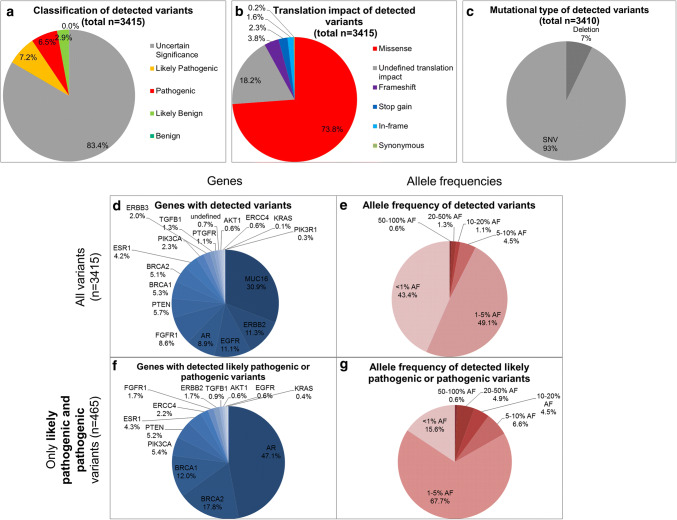
Fig. 2.
Characteristics of called variants in cfDNA in HR+ HER2– MBC patients (n= 40).a Classification of variants according to their known impact (benign, likely benign, uncertain significance, likely pathogenic and pathogenic) [26] done by IVA. Of all detected variants (n = 3415), 14% are likely pathogenic or pathogenic. b Translation impact of detected variants. Three quarter of all variants were missense variants. c Differentiation into mutational types. Only n = 5 were multi nucleotide polymorphisms or insertions, while 93% were single nucleotide polymorphisms. d Distribution of variants regarding their gene location. Most variants were found in the MUC16 gene. e AFs of all variants. 90% of all variants displayed AFs < 5%. f Distribution of all pathogenic or likely pathogenic variants (n = 465) regarding their gene location. Most pathogenic or likely pathogenic variants found in the AR gene. g AFs of all pathogenic and likely pathogenic variants. 68% presented an AF between 1 and 5%