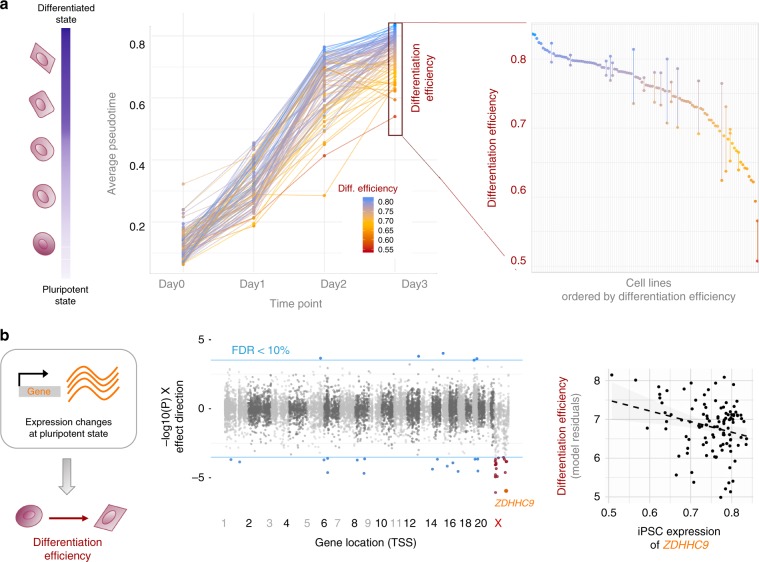
Fig. 5. Identification of molecular markers for differentiation efficiency.
a Variation in differentiation efficiency across cell lines. Left: Differentiation progress over time, showing trajectories for 98 cell lines, coloured by differentiation efficiency. Shown are 98 cell lines with sufficient data at all time points (out of 126, more than 10 cells). Differentiation efficiency of a cell line was defined as the average pseudotime across all cells on day 3. Right: Differentiation efficiency across cell lines (points), and consistency of individual cell lines differentiated in multiple experiments (vertical bars). b Associations between iPSC gene expression levels and differentiation efficiency. Left: schematic. Centre: Genome-wide analysis to identify markers of differentiation efficiency, considering iPSC gene expression levels. Displayed are negative log P-values signed by the direction of the effect. Horizontal blue lines denote FDR = 10% (Benjamini-Hochberg adjusted). Autosomal genes with significant associations are shown in blue; X chromosome genes with significant associations are shown in red. Right: Scatter plot between gene expression in the iPS state and differentiation efficiency for the X chromosome gene ZDHHC9.