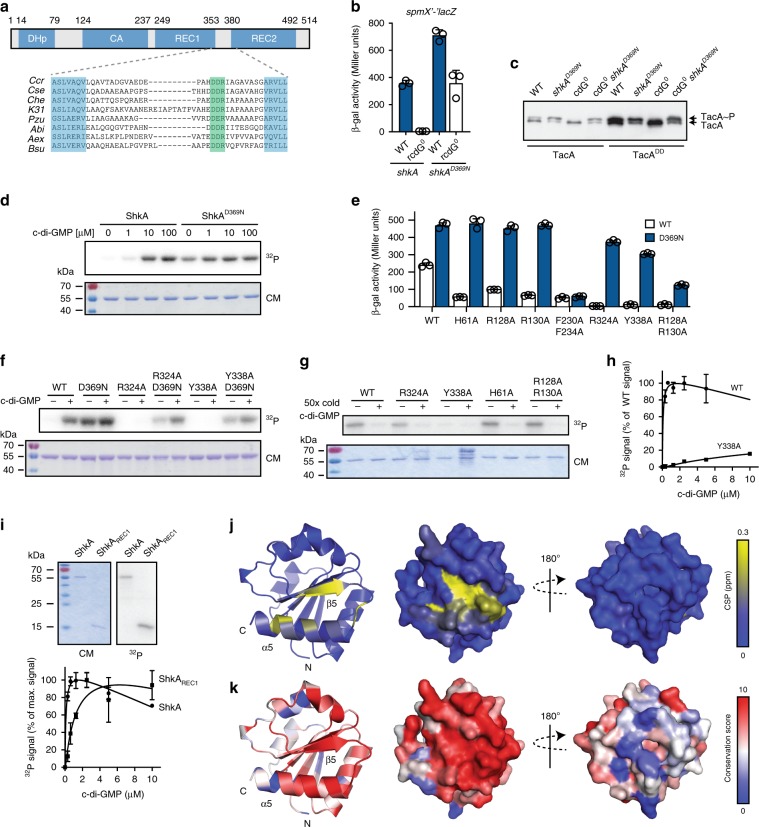
Fig. 2. C-di-GMP binds to the REC1 pseudo-receiver domain.
a ShkA domain architecture drawn to scale (top) and alignment of the REC1-REC2 linker harboring the DDR motif (highlighted in green) of ShkA orthologs (bottom). Ccr, C. crescentus; Cse, Caulobacter segnis; Che, Caulobacter henricii; K31, Caulobacter sp. K31; Pzu, Phenylobacterium zucineum; Abi, Asticcacaulis biprosthecium; Aex, Asticcaucaulis excentricus; Bsu, Brevundimonas subvibrioides. Dimerization and histidine-phosphotransfer (DHp), catalytic and ATP-binding (CA) and receiver (REC1, REC2) domains are indicated. b Activity of the spmX promoter in indicated strains harboring plasmid pAK502-spmX. shkAD369N is expressed from the native chromosomal locus. Means and standard deviations are shown (N = 3). c Phos-tag PAGE immunoblots of indicated strains producing 3xFLAG-tagged TacA or TacADD from the native locus. d In vitro autophosphorylation assays of ShkA and ShkAD369N. Top: autoradiograph; bottom: Coomassie stain. e β-Gal assays of ΔshkA mutant strains harboring plasmid pAK502-spmX and expressing different shkA alleles in trans from plasmid pQF with the indicated amino acid substitutions alone (WT, white bars) or in combination with D369N (blue bars). Shown are means and standard deviations (N = 3). f In vitro autophosphorylation assays of wild type or mutant ShkA with (10 µM) or without c-di-GMP. Reactions were run for 5 min. Top: autoradiograph; bottom: Coomassie stain. g Autoradiographs of purified ShkA and ShkA mutant variants (0.5 µM) UV-crosslinked with 10 µM [32P]c-di-GMP with or without addition of a 50-fold molar excess of non-labeled c-di-GMP. Top: autoradiograph; bottom: Coomassie stain. h Quantified autoradiographs of purified ShkA and ShkAY338A (0.5 µM) UV-crosslinked with increasing concentrations of [32 P]c-di-GMP. Shown are mean values and standard deviations (N = 2). i Autoradiographs and Coomassie stain of the same gel of purified ShkA and the isolated REC1 domain (ShkAREC1) (0.5 µM) after UV crosslinking with 10 µM [32P]c-di-GMP (top). Quantified autoradiographs of purified ShkA and ShkAREC1 (0.5 µM) after UV crosslinking with increasing concentrations of [32P]c-di-GMP (bottom). Means and standard deviations are shown (N = 2). j Cartoon and surface representation of the ShkAREC1 homology model with NMR chemical shift perturbations (CSPs) upon c-di-GMP binding indicated by a blue-to-yellow gradient. k Conservation score of ShkA orthologs (see “Methods”). Source data are provided as a Source Data file.