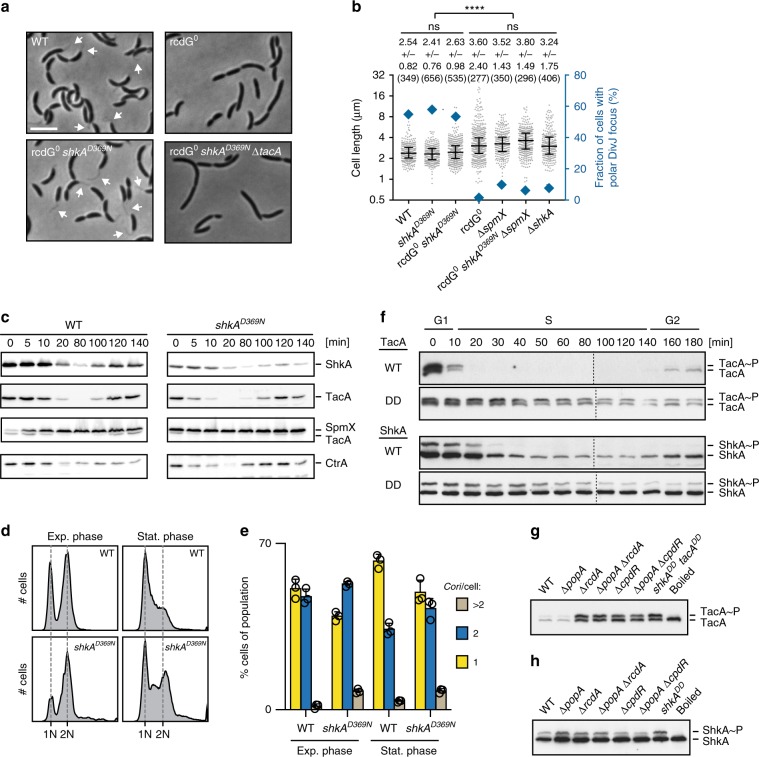
Fig. 3. C-di-GMP imposes precise temporal control of TacA activity during G1/S.
a Representative phase-contrast micrographs of indicated strains. Arrows point to stalks. The scale bar represents 4 µm. b Quantification of cell length and polar DivJ localization (DivJ-mCherry) of indicated strains. Median values with interquartile ranges are shown in the graph and mean values and standard deviations are indicated above the graph. The number of cells analyzed is shown in brackets. **** indicates a P value of < 0.0001; ns, not significant (ordinary one-way ANOVA and Tukey’s multiple comparison test). c Immunoblots of synchronized cultures of C. crescentus strains expressing 3xFLAG-shkA or 3xFLAG-shkAD369N from the native chromosomal locus were probed with anti-FLAG, anti-TacA, anti-SpmX and anti-CtrA antibodies. d Analysis of chromosome content by flow cytometry of indicated strains in exponential or stationary phase after rifampicin treatment. e Quantification of chromosome number of indicated strains. Shown are means and standard deviations (N = 3). f Phos-tag PAGE immunoblots of synchronized cultures of strains expressing chromosomally encoded 3xFLAG-tacA, 3xFLAG-shkA, 3xFLAG-tacADD, or 3xFLAG-shkADD alleles were probed with anti-FLAG antibody. g Phos-tag PAGE immunoblots of mixed cultures of indicated mutant strains expressing 3xFLAG-tacA probed with anti-FLAG antibodies. h Phos-tag PAGE immunoblots of mixed cultures of indicated mutant strains expressing 3xFLAG-shkA probed with anti-FLAG antibodies. Source data are provided as a Source Data file.