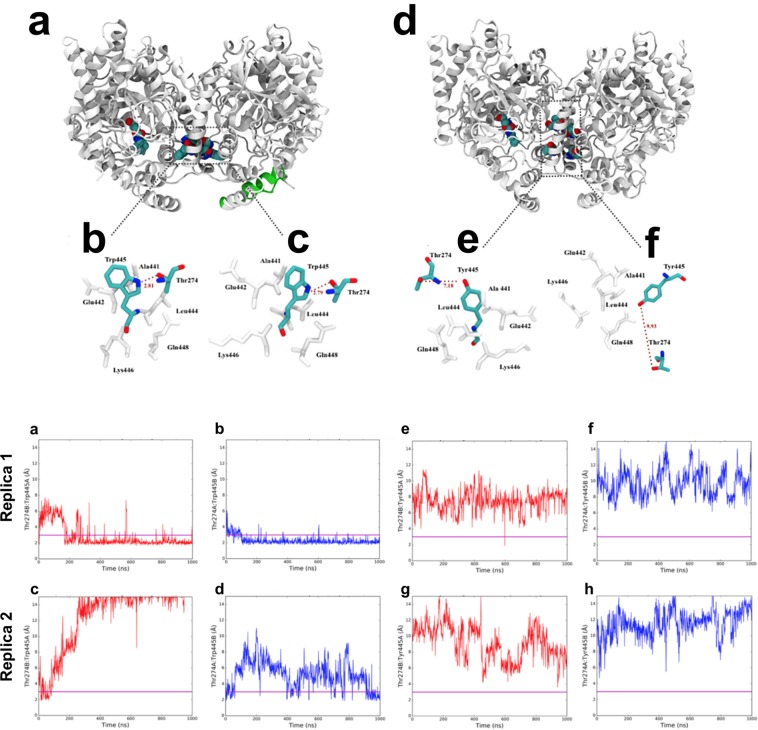
Figure 3.
Molecular dynamics of rFAAH and its W445Y mutant. Upper Panel: Last frame snaphots of MD simulations of the URB597/rFAAH (a) and the URB597/W445Y-rFAAH complexes (d), where the URB597 molecule is covalently bound in the active site of one monomer only (monomer A). Schematic representation of most frequent W445:T274 interactions (b,c) and Y445:T274 interactions (e,f) over the simulation is reported; related residues are also depicted in the dotted squares (a,d). Distances between residues are indicated with red broken lines (b,c,e,f). Lower Panel: The HB plots URB/rFAAH complex (a–d), where distances (expressed in Å) between the OH group of T274 and the W445’s sidechain nitrogen over all the trajectory length, are shown. These simulations indicate that the T274 of monomer A of wild type rFAAH establishes a hydrogen bond with the W445 of monomer B (and viceversa) in 3 cases of the 4 analyzed trajectories length. The HB Plots of URB/W445Y-rFAAH complex (e–h) show that in the rFAAH mutant the distances between the OH group of T274 and Y445’s sidechain hydroxyl in both monomers and all replicas (e–h) are not compatible with the formation of a hydrogen bond. These MD simulations suggest that the hydrogen bond is more likely to form in the wild-type rFAAH. A line y = 3 highlights peaks over 3 Å for each of the plot.