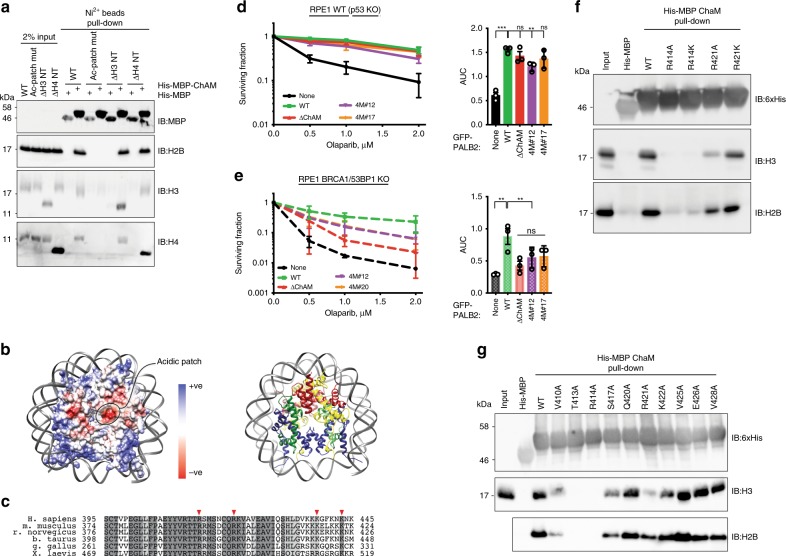
Fig. 4. ChAM domain of PALB2 interacts with nucleosome acidic patch.
a In vitro pull-down (PD) assay using purified components. 6xHis-MBP-tagged PALB2 ChAM domain or 6xHis-MBP immobilised on Ni-NTA affinity beads, incubated with recombinant nucleosome variants. Acidic patch mutant (Ac-patch mut) comprises H2A E61/91/92A and H2B E105A; ΔH3 NT comprises H3 residues 25−135; ΔH4 NT comprises H4 residues 21−106. Anti-H3 and H4 antibodies showed increased reactivity towards histones with shorter N-terminal tails. b The nucleosome and its acidic patch. Left panel: electrostatic potential view of the protein histone surface; right panel: ribbon representation of the nucleosome core particle. c Sequence alignment of ChAM with alanine scanning mutation areas highlighted (blue line) and basic residues mutated in 4 M mutant highlighted (red arrows). d, e Clonogenic survivals of RPE1 p53 KO (e) or p53/BRCA1/53BP1 KO (f) cells complemented with PALB2WT, PALB2∆ChAM or PALB24M in response to indicated doses of olaparib. Lower panels show corresponding area under curve (AUC) bar graphs; n ≥ 3. The bars represent mean ± s.e.m.; one-way ANOVA; **P < 0.01, ***P < 0.001; ns, not significant (P ≥ 0.05); n = 3 independent experiments. Source data are provided as a Source Data file. f Pull-down assays comparing NCP association of 6xHis-MBP-PALB2 ChAM variants with charge removal and charge retaining ChAM mutations on Arg414 and Arg421. g Pull-down assays between NCPs and immobilised 6xHis-MBP-ChAM variants containing alanine mutations across the proposed nucleosome-interacting region.