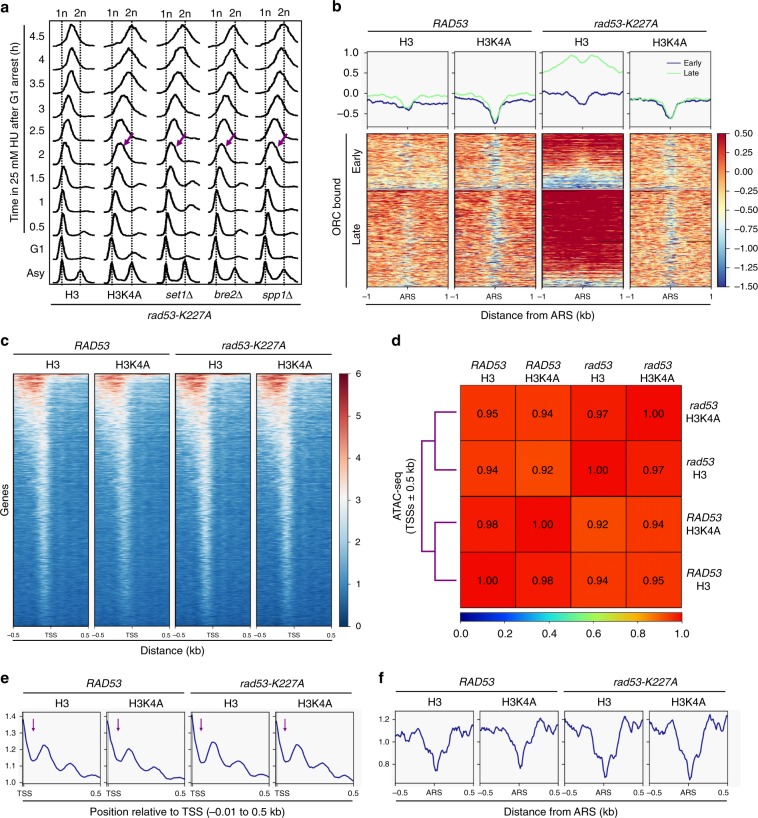
Fig. 3. H3K4 methylation causes fork stalling in rad53 mutants following replication stress without altering chromatin structure.
a Cells with H3, H3K4A, set1Δ, bre2Δ, and spp1Δ on the rad53-K227A background were arrested in G1 and released into 25 mM of HU for 2 h. The cellular DNA content was determined by FACS at the indicated time-points. Purple slanted arrows indicate faster replication progression in mutant cells compared to H3 cells. b The graphs show DNA Pol2 binding profiles centered on ORC-bound early or late ARSs in RAD53-H3, RAD53-H3K4A, rad53-K227A-H3, and rad53-K227A-H3K4A cells. c ATAC-seq signals on TSSs ± 0.5 kb regions after G1-release in 25 mM HU. d Spearman’s rank correlation coefficient of ATAC-seq signals according c. e ATAC-seq signals on TSS regions (−0.1 to 0.5 kb). f ATAC-seq signals on ORC-bound origins.