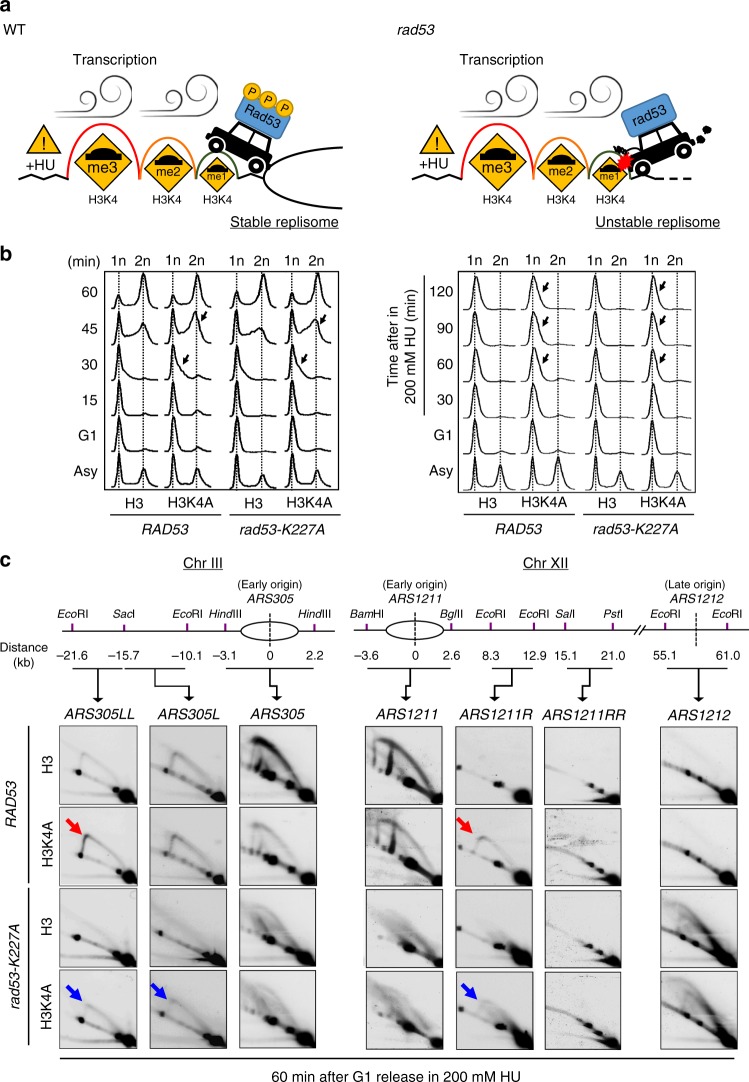
Fig. 6. H3K4 methylation deposited by transcription decelerates replication forks.
a A graphic depiction of the “speed bump model”. Sites of H3K4 methylation deposited by transcriptional activity act as “speed bumps” for active replication forks (represented by the black car), while activation of Rad53 checkpoint kinase maintains the stability of replisomes during replication stress. In the absence of checkpoint activity, such as in rad53-mutant cells, H3K4 methylation becomes an impediment to replication and destabilizes the replisome. The red explosion drawing indicates the stalling of replication forks. b FACS profiles of RAD53-H3, RAD53-H3K4A, rad53-K227A-H3, and rad53-K227A-H3K4A cells released from G1 into rich medium without (left) or with 200 mM HU (right) at indicated time points. Black arrows indicate faster S-phase progression or DNA duplication in H3K4A cells compared to H3 cells. c 2D gel analysis of RAD53-H3, RAD53-H3K4A, rad53-K227A-H3, and rad53-K227A-H3K4A cells. Replication intermediates from indicated regions of chromosomes III and XII from ARS305 and ASR1211 were identified in the indicated strains following G1 synchronization and release into 200 mM HU for 1 h. The arrows mark Y-arc signals that represent duplicating replication forks in RAD53-H3K4A and rad53K227A-H3K4A cells. Bubble-arc signals at ARS1212 indicate the late origin was repressed in RAD53 and activated in rad53-K227A cells.