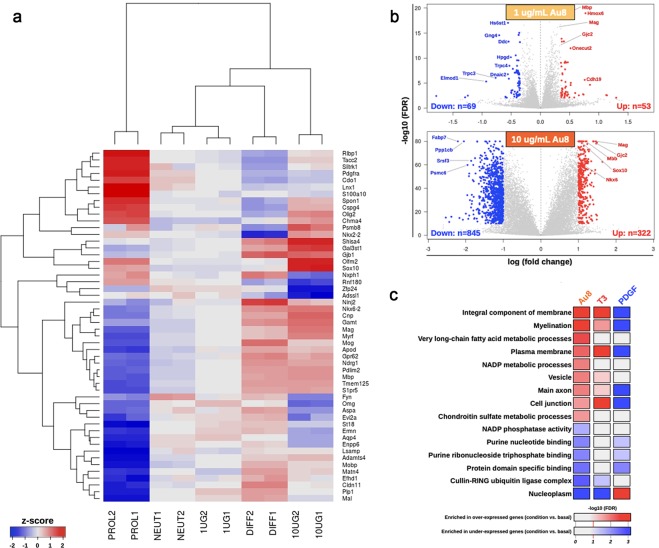
Figure 7.
Transcriptomics analyses identified OL differentiation and myelination pathways that are upregulated in response to CNM-Au8. Isolated OPC cultures were treated with vehicle (NEUT1 and NEUT2), 1.0 (1UG1 and 1UG2) or 10.0 μg/mL (10UG1 and 10UG2) CNM-Au8, 0.01 μg/mL PDGF to induce proliferation (PROL1 and PROL2), or 0.04 μg/mL T3 (DIFF1 and DIFF2) to induce differentiation, for 72 h in duplicate. Cells were then processed for RNAseq analyses. (a) The top fifty variable genes in the dataset are displayed in the heatmap, with the name of each gene in each row labeled to the right. Samples were hierarchically clustered (dendrogram at top), demonstrating that CNM-Au8 treated cells’ transcript profiles are closer, but not overlapping, that of T3-treated cells, as compared to PDGF treated cells. (b) Volcano plots of differentially expressed genes, with the identities of upregulated genes known to play a role in differentiation and myelination indicated in red. (c), Gene ontology enrichment analysis revealed that gene pathways associated with myelination, plasma membrane regulation, and fatty acid metabolism were among those upregulated upon treatment with CNM-Au8, as well as, in part, with T3.