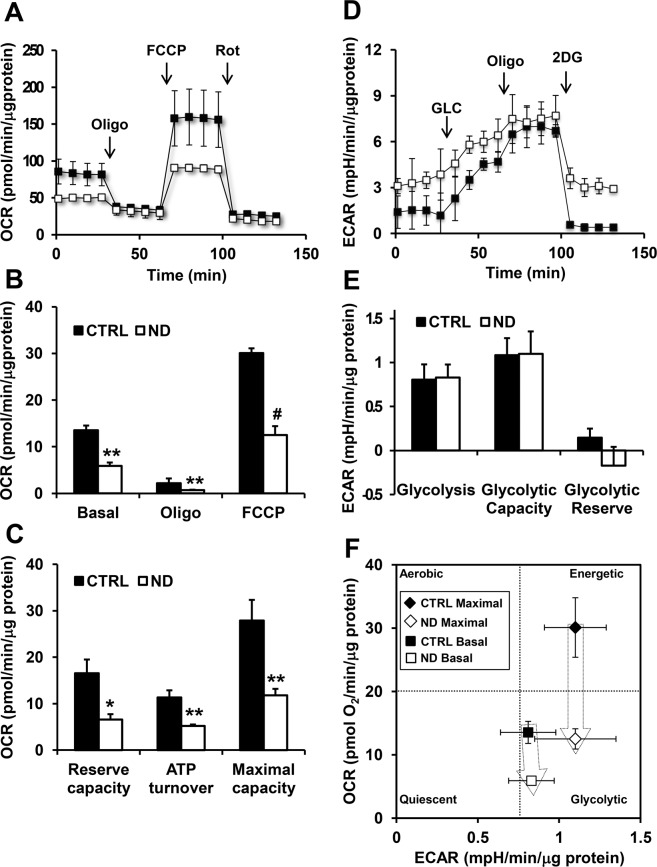
Figure 3.
Metabolic flux analysis of Nandrolone-treated HepG2 cells. Representative oxygen consumption rates (OCR) (A) and extra cellular acidification rates (ECAR) (D) profile of HepG2 cells treated with vehicle (CTRL) or 80 μM nandrolone (ND), assayed by the Seahorse XFe96 Analyzer as described under Materials and Methods. Oligomycin (Oligo), FCCP, and Rotenone (Rot) were added at the indicated points in A. Glucose (GLC), Oligo and 2-deoxyglucose (2DG) were added at the indicated points in D. The histograms in B,E, show OCR and ECAR normalized to the protein content of the cells removed from each well at the end of the assay. OCR in B Basal, resting OCR; Oligo, OCR measured after the addition of the ATP synthase inhibitor oligomycin also referred as proton leak; FCCP, OCR measured after the addition of the uncoupler FCCP eliciting the maximal respiratory capacity. The OCR were corrected for the residual OCR measured after the addition of the CxI inhibitor rotenone (not shown). ECAR in E Glycolysis, resting ECAR, measured after the addition of glucose and corrected for the 2DG-insensitive ECAR; Glycolytic Capacity, ECAR measured after the addition of oligomycin and refers to the maximal glycolytic activity with the OxPhos inhibited; Glycolytic Reserve, difference between ECAR measured in the presence of oligomycin and under resting conditions. (C) The histograms show Reserve Capacity, ATP turnover and maximal capacity computed as described in Materials and Methods. (F) Energy map obtained plotting the basal and maximal OCR and basal and maximal ECAR measured in A,D The values are means ± SEM of three independent experiment carried out in 3 technical replicates under each condition; *P < 0.05; **P < 0.01; #P < 0.001.