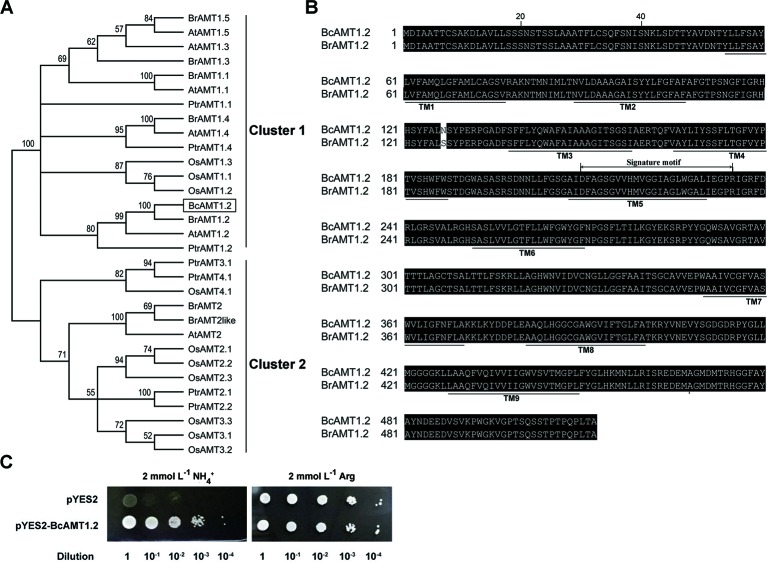
Figure 5.
Sequence analysis of BcAMT1.2 and functional complementation in yeast mutant 31019b cells by BcAMT1.2. (A) Phylogenetic tree of AMT homologs. It was constructed by the Neighbor-Joining method in MEGA 6.0. Bootstrap values were derived from 1000 replications, and evolutionary distances were estimated in terms of the number of amino acid substitutions per site. The numbers at the nodes are bootstrap values. Accession numbers of protein sequences of AMTs from given plant species are listed in Supplementary Table S2 . At, Arabidopsis thaliana; Bc, Brassica campestris; Br, Brassica rapa; Os, Oryza sativa; Ptr, Populus trichocarpa. BcAMT1.2 was represented by a black box. (B) Amino acid sequence alignment of AMT1.2 from B. campestris and B. rapa. The alignment was performed using ClustalW. Amino acids are presented as capital letters; residues are shown in white letters on black if two sequences have identical residues at the aligned positions. Thick lines below sequences show the positions of potential transmembrane α-helices (TMs) as predicted using Protter (http://wlab.ethz.ch/protter/). The sequences marked signature motif indicated a motif specific to the AMT1 sub-family identified using Weblogo (http://weblogo.berkeley.edu/logo.cgi/). (C) Functional complementation in yeast mutant 31019b cells by BcAMT1.2. pYES2: empty vector was used as negative control, pYES2-BcAMT1.2: BcAMT1.2 ORFs was cloned into pYES2 vector. Yeast cell suspensions were adjusted to an optical density at 600 nm of 1.0 (dilution 1), and serially diluted by factors of 10. For each dilution, 3 μL of the yeast cell suspensions were spotted on yeast N base medium with 2 mmol L–1 NH4 + or arginine.