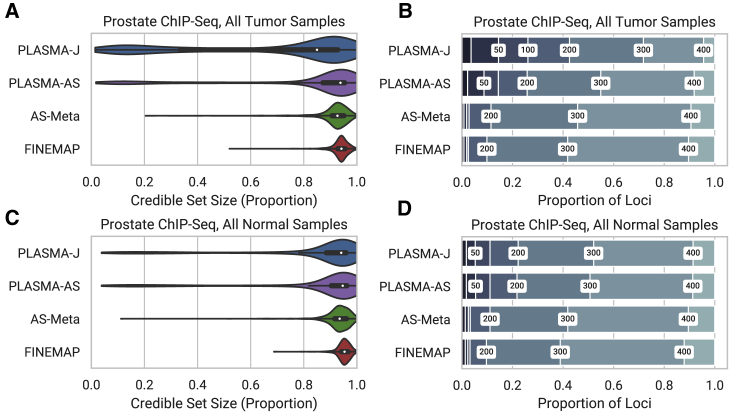
Figure 7.
Comparison of 95% Confidence Credible Set Sizes for Peaks in Prostate Tumor and Normal Cells, with an Allelic Imbalance False Discovery Rate of 0.05
Presence of H3k27ac histone marks was quantified as ChIP-seq read counts mapped to phased genotypes.
(A and C) Distribution of credible set sizes for tumor and normal samples, respectively, under a 1-causal-variant assumption.
(B and D) Proportion of peaks fine mapped to 95% credible sets within given thresholds. Thresholds used were 20, 50, 100, 200, 300, and 400.