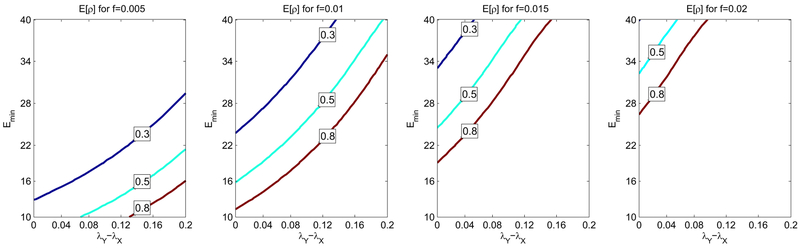
Fig. 5.
Contour lines of the expected prevalence of mixed infection E[ρ] are drawn for four different values of the sensitivity threshold f of the genotyping method. Every plot shows three elevation levels (0.3, 0.5 and 0.8) when the difference of the growth rates λy – λX spans between 0 and 0.2 (x-axis) and the expected number of minority strain cells in sputum Emin spans between 10 and 40 (y-axis). To produce each plot we simulated a study involving 500 patients among whom 75 are detected with mixed infection. From the comparison of the contour plot we evince that as the threshold f increases, a greater portion of the parameter space features a high (> 0.8) expected prevalence of mixed infection. On the other hand, when f is small, E[ρ] is closer to the detected prevalence 15%. This not only confirms that a small sensitivity threshold allows more precise results, but also shows that even when such threshold is small, the raw percentage 15% should be corrected to give a good estimate of the real prevalence of mixed infection.