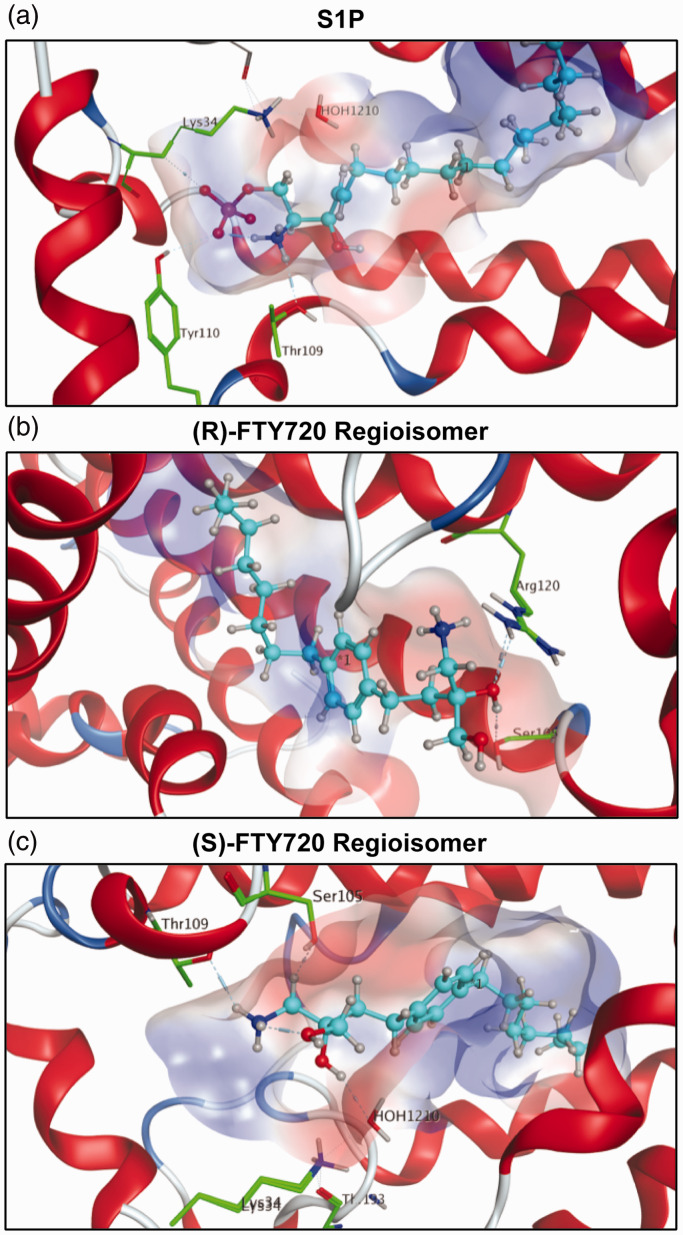
Fig. 4.
S1P1 docking simulations with S1P and (S)- and (R)-FTY720 regioisomers. In all visualizations, the substrate is highlighted in light blue. The polarity of the active site’s cavity is indicated by colors: red regions define hydrophilicity, blue areas mark lipophilicity. Receptor residues engaging in interactions with the substrate are highlighted in green with emphasis of respective hydrogen bonds. (a) S1P in its most stable conformer within the active site of S1P1 with a calculated relative energy of −351.72 kcal/mol. The phosphate moiety of S1P is largely responsible for the strong binding, as one of the negatively charged oxygens of the phosphoester acts as a hydrogen bond acceptor for residue Tyr110, and one forms a hydrogen bond with residue Lys34. The protonated amino group of S1P donates to another hydrogen bond with Thr109 and forms an intramolecular hydrogen bond with the phosphate oxygen that binds to Tyr110. The calculations suggest that residues Lys34, Thr109, and Tyr110 lock the hydrophilic phosphate head group of S1P into position, while the hydrophobic tail of S1P is held between the seven lipophilic transmembrane helices of S1P1, well isolated from polar media. The hydroxyl group at the C3 position fails to display interactions with the active site of S1P1. (b) The unphosphorylated FTY720 regioisomer 3R is depicted in the active site of S1P1 in its energetically most favorable conformer, its energy being calculated as −1.92 kcal/mol, indicating no favorable S1P1 binding. Aside from the substrate’s C3 hydroxyl group forming hydrogen bonds with residues Ser105 and Arg120, the docking simulation did not show any other interactions. (c) The unphosphorylated FTY720 regioisomer 3S is depicted in its calculated energetically most stable conformation in the active site of S1P1 with a relative energetic value of −0.81 kcal/mol, which also suggests poor binding to S1P1 in comparison to S1P. The docking simulation of unphosphorylated 3S to S1P1 shows hydrogen binding of the protonated amine of 3S to residue Thr109. In the calculations, the hydroxyl group at C3 further coordinates to a water molecule which in turn hydrogen bonds to residue Lys34. No direct interaction of 3S with any residues except for Thr109 could be observed in the calculations, explaining the poor affinity of the unphosphorylated substrate to S1P1.
S1P: sphingosine-1-phosphate; FTY720: 2-amino-2-(2-[4-octylphenyl]ethyl)-1,3-propanediol.