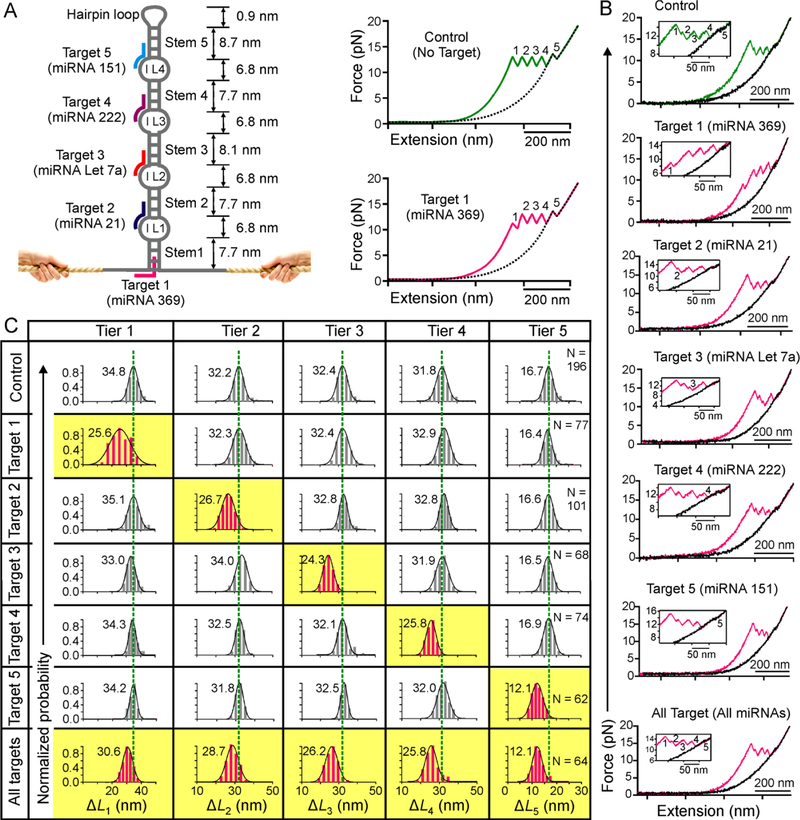
Figure 5.
Single-molecule probe capable of large-scale topochemical analyses of miRNAs. (A) Schematic of the hairpin sensor that contains four internal loops (IL1-IL4) and 5 stems. Target 1 (pink) binds with the toehold at the base of the probe and invades into stem 1. Targets 2–4 bind with internal loop toeholds and half-length of the corresponding stems. Without target, the DNA construct produces five well addressed unfolding features (top right). With target 1, both RF and ΔL of the first feature are expected to be smaller (bottom right). Dotted curves depict relaxing F-X traces. Note feature 5 indicates the transition for the stem linked to the hairpin tetraloop. (B) Representative F-X curves of the probe without (green) and with (pink) 50 nM miRNAs in 10 mM Tris buffer (pH 7.4 with 100 mM KCl). (C) Change-in-contour-length (ΔL) histograms of a particular unfolding feature without (grey) and with (pink) 50 nM miRNA targets. Dotted green lines are the guides to the eye.