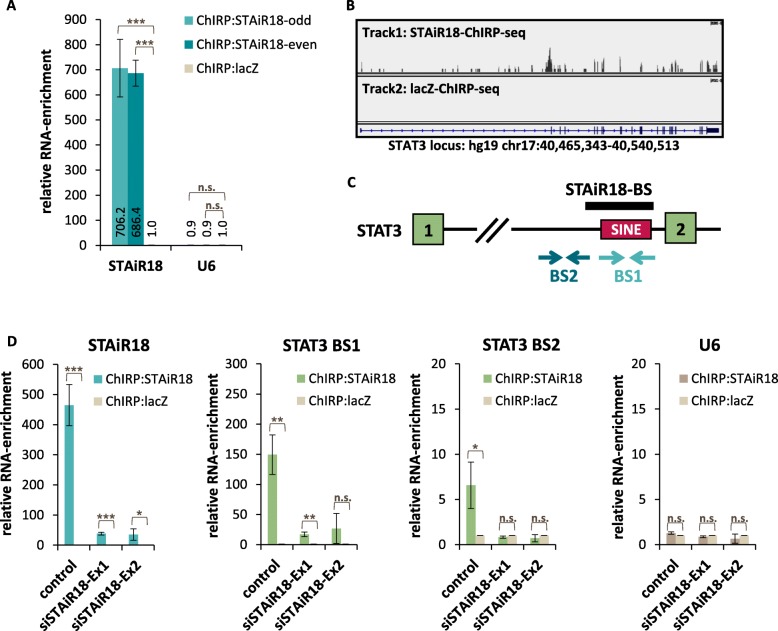
Fig. 5.
STAiR18 interacts with STAT3 mRNA. a STAiR18 ChIRP is highly specific and efficient. Pulldown of STAiR18-RNA from crosslinked INA-6 cells was realized by oligos covering exon 1 and 2. Oligos were then divided into two pools (odd and even numbered). For control, oligos targeting lacZ RNA were used. After pulldown, RNA was analyzed by qPCR (n = 4). b STAiR18 ChIRP followed by RNA-sequencing revealed an association of STAiR18 with STAT3 on RNA-level. Shown is the IGV window of the STAT3 locus after STAiR18 (Track1 with STAiR18 binding peak) and lacZ (Track2) ChIRP-seq. c The STAT3 gene harbors a SINE within the first intron corresponding to the binding site (BS) of STAiR18. Binding sites for primers marking the specific STAiR18-BS1 are shown in light blue, a primer pair for a upstream region BS2 is shown in dark blue. d Specific interaction of STAiR18 with STAT3 RNA was determined in INA-6 cells after KD of STAiR18 exon1 or 2, followed by a ChIRP experiment using oligonucleotides targeting STAiR18 or lacZ 24 h post-transfection. RNA was analyzed by qPCR using specific primers targeting the STAiR18-BS within the STAT3 intron. Primer pairs for STAiR18 were used as positive, for U6 as negative control. Values were normalized to lacZ (n = 3)