Figure 3.
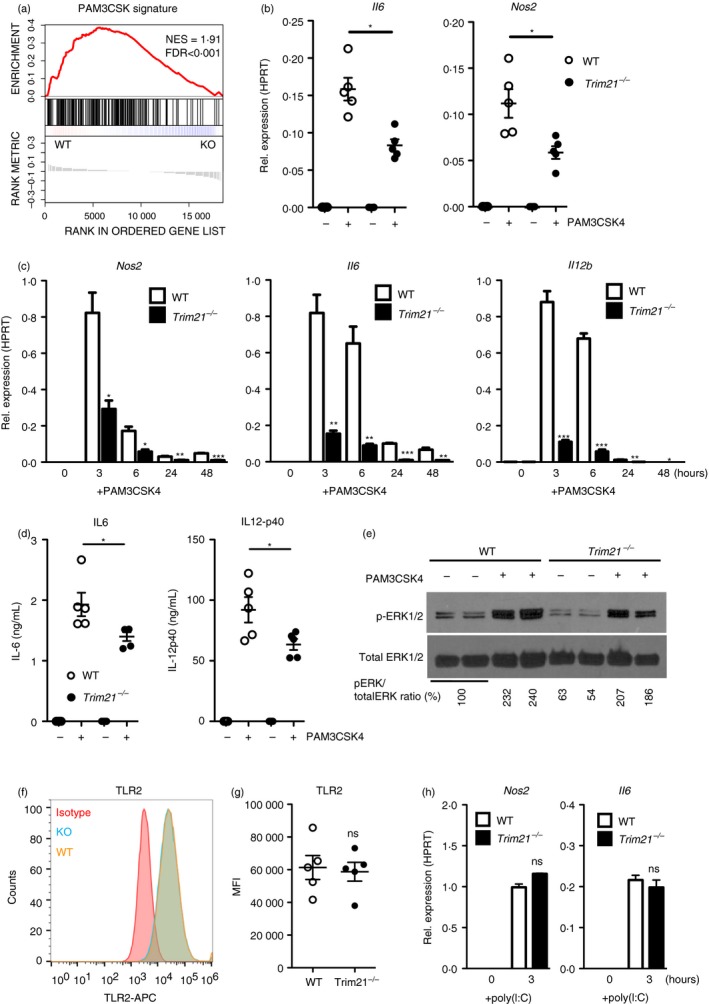
Trim21 −/− bone‐marrow‐derived macrophages (BMDMs) are hyporesponsive to stimulation with PAM3CSK4. Gene‐set enrichment analysis of Toll‐like receptor 2 (TLR2) ‐activation (PAM3CSK4) signature genes in Trim21 −/− (knockout; KO) and wild‐type (WT) BMDMs at baseline (a). Trim21 −/− and WT BMDMs were stimulated with 0·1 µg/ml PAM3CSK4 for 6 hr followed by quantitative RT‐PCR against Nos2 (iNOS) and Il6 (IL‐6) (n = 5) (b). Trim21 −/− and WT BMDMs were stimulated with 0·1 µg/ml PAM3CSK4 for 3–48 hr followed by quantitative RT‐PCR against Nos2 (iNOS), Il6 (IL‐6) and Il12b (IL‐12‐p40) (n = 3) (c). Trim21 −/− and WT BMDMs were stimulated with 0·1 µg/ml PAM3CSK4 for 24 hr, followed by ELISA to measure secreted IL‐6 and IL‐12‐p40 protein (n = 5) (d). Immunoblotting against phosphorylated EKR1/2 (p‐ERK1/2) and total ERK1/2 in BMDMs stimulated with 0·1 µg/ml PAM3CSK4 for 6 hr (e). Trim21 −/− and WT BMDMs were stimulated with 1·0 µg/ml poly(I:C) for 3 hr followed by quantitative RT‐PCR against Nos2 and Il6 (n = 3) (f). Surface expression of TLR2 on BMDMs was determined by flow cytometry (n = 5) (g, h). Statistics for gene expression data were calculated using the Student's t‐test, and statistical testing for flow cytometry data was performed using two‐sided Mann–Whitney test (*P < 0·05, **P < 0·01, ***P < 0·005). The experiment was performed three times with similar results. Gene‐set enrichment was calculated as a normalized enrichment score. Multiple testing was corrected using false‐discovery rate calculated based on gene‐set permutations.
