Abstract
Background
Primary vaginal melanomas are uncommon and aggressive tumors with poor prognosis, and the development of new targeted therapies is essential. This study aimed to identify the molecular markers occurring in these patients and potentially improve treatment strategies.
Materials and Methods
The clinicopathological characteristics of 36 patients with primary vaginal melanomas were reviewed. Oncogenic mutations in BRAF, KIT, NRAS, GNAQ and GNA11 and the promoter region of telomerase reverse transcriptase (TERT) were investigated using the Sanger sequencing. The expression and copy number of programmed death‐ligand 1 (PD‐L1) were also assessed.
Results
Mutations in NRAS, KIT, and TERT promoter were identified in 13.9% (5/36), 2.9% (1/34), and 5.6% (2/36) of the primary vaginal melanomas, respectively. PD‐L1 expression and amplification were observed in 27.8% (10/36) and 5.6% (2/36) of cases, respectively. PD‐L1 positive expression and/or amplification was associated with older patients (p = .008). Patients who had NRAS mutations had a poorer overall survival compared with those with a wild‐type NRAS (33.5 vs. 14.0 months; hazard ratio [HR], 3.09; 95% CI, 1.08–8.83). Strikingly, two patients with/without PD‐L1 expression receiving immune checkpoint inhibitors had a satisfying outcome. Multivariate analysis demonstrated that >10 mitoses per mm2 (HR, 2.96; 95% CI, 1.03–8.51) was an independent prognostic factor.
Conclusions
NRAS mutations and PD‐L1 expression were most prevalent in our cohort of primary vaginal melanomas and can be potentially considered as therapeutic targets.
Implications for Practice
This study used the Sanger sequencing, immunohistochemistry, and fluorescence in situ hybridization methods to detect common genetic mutations and PD‐L1 expression and copy number in 36 primary vaginal melanomas. NRAS mutations and PD‐L1 expression were the most prevalent, but KIT and TERT mutations occurred at a lower occurrence in this rare malignancy. Two patients receiving immune checkpoint inhibitors had a satisfying outcome, signifying that the PD‐L1 expression and amplification can be a possible predictive marker of clinical response. This study highlights the possible prospects of biomarkers that can be used for patient selection in clinical trials involving treatments with novel targeted therapies based on these molecular aberrations.
Keywords: Primary vaginal melanoma, Ooncogenic mutations, NRAS, PD‐L1 expression
Short abstract
Little is known about the molecular characteristics of primary vaginal melanoma. This article reports on the molecular markers of this rare and aggressive disease, focusing on improvements in treatment strategies.
Introduction
The incidence of melanoma is on a gradual rise and growing at a faster rate than other solid tumors 1. Vaginal malignant melanoma, one of its highly aggressive subtypes and often diagnosed at an advanced stage, is an extremely rare mucosal melanoma that accounts for 2.4%–2.8% of all vaginal cancers and 0.3%–0.8% of all malignant melanomas 2. Nowadays, there are multiple therapeutic strategies for treating vaginal melanomas, including surgery, radiotherapy, chemotherapy, targeted therapy, boron neutron capture therapy, and immunotherapy, but their prognoses are still dismal, with a 5‐year survival rate ranging from 0% to 32.3% 3, 4, 5, 6, 7, 8.
In‐depth research on melanoma has shown that targeted and immunotherapies could improve the outcomes in patients with melanoma 9. Previous studies have shown that mucosal melanomas at different anatomic locations exhibit different oncogenic aberrations 10, 11, 12, 13. Several studies have focused on identifying the molecular alterations of melanomas composed of specific mutations in NRAS, BRAF, KIT, TERT, and GNAQ/GNA11, observed in distinct subtypes of melanomas. The overall rate of BRAF mutation in melanoma has been found to be up to 67% 14, 15, and the mutation frequently occurs in non‐chronically sun‐damaged (CSD) skin. NRAS is mutated in 10%–25% of cutaneous melanomas and occurs most frequently at hotspots in codons 12 and 61 16, 17, 18 and activates downstream effectors. An increase in copy number (up to 25%) and mutations (10%–20%) of KIT in mucosal, acral, and CSD melanomas were identified 19. Mutations in GNAQ/GNA11, a gene encoding an α subunit of heterotrimeric G proteins, are found in up to 83% of uveal melanomas 20, 21, 22. GNAQ and GNA11 mutations in melanomas affect codons 209 or 183 and result in consistent activation of the protein kinase C and MAPK pathways 21, 23. TERT, which encodes the catalytic subunit of telomerase, is mutated in cutaneous melanoma 24, 25, 26 and has been found to be associated with aggressive behavior of the melanoma and a poorer prognosis 25, 27. In recent years, mutant‐selective BRAF 28, MEK 29, 30, 31, and KIT inhibitors 32 have demonstrated impressive clinical results in molecularly selected patients.
Several previous studies have demonstrated that patients wth melanoma, non‐small cell lung cancer, and renal cell carcinoma could achieve a 10%–40% clinical response with immune checkpoint inhibitions 33, 34. However, approximately 7%–34% of these cases do also experience high‐grade immune‐related adverse events 35, 36. Therefore, to increase treatment compliance and outcome, appropriate biomarkers capable of predicting response are highly needed for identifying patients who would be most beneficial to these targeted therapies. Of them, the programmed death‐ligand 1 (PD‐L1) is the most broadly investigated and binds to inhibitory checkpoint molecule PD‐1. The detection of PD‐L1 expression in tumor cells or tumor‐associated stromal cells by immunohistochemistry (IHC) has enabled the identification of tumors which would response to anti‐PD‐L1 blockade 34, 37, 38. However, published correlative data for vaginal melanoma remain scarce.
In the present study, we performed an analysis of the clinicopathological features of 36 patients with primary vaginal melanoma in a single institution. Further major molecular alterations including the PD‐L1 status were characterized to improve the current understanding of altered molecular pathways and thereby explore possible strategies for their therapeutic management.
Subjects, Materials, and Methods
Study Participants
A total of 36 primary vaginal melanomas samples were collected from patients treated at the Sun Yat‐sen University Cancer Center between March 2004 and February 2018. Of them, 32 had surgery as their primary treatment, including radical surgery and local excision with wide margin, 2 were treated with chemoradiotherapy or immune checkpoint inhibitors after biopsy, and 2 refused treatment after diagnosis. Of those 32 patients, 20 received chemotherapy, 12 received radiotherapy, 5 received a second‐time surgical resection, 2 received interferon‐α, and 1 received immune checkpoint inhibitors during the course of subsequent treatment (supplemental online Table 1). The following pathological characteristics of tumor were evaluated: presence or absence of ulceration or pigmentation, depth of invasion (DOI; measured from the outermost point of the mucosa to the deepest point of invasion), number of mitoses per mm2, and the predominant cell type (epithelioid, spindle cell, or mixed). The tumor was staged according to the 8th edition of the American Joint Committee on Cancer staging system for vaginal melanoma 39. The protocol was designed in accordance with the Declaration of Helsinki. This study was approved by the Research Ethics Committee of the Sun Yat‐sen University Cancer Center (No. B2016–069–01).
DNA Isolation and Genetic Mutation Detection
Formalin‐fixed, paraffin‐embedded (FFPE) tissue blocks were reviewed for quality control, and the regions containing more than 50% of tumor cells were selected for macrodissection. Genomic DNA was extracted using the QIAamp DNA FFPE Tissue kit (QIAGEN, Hilden, Germany). Direct sequencing of KIT (exons 9, 11, 13, 17, and 18), NRAS (exons 2 and 3), BRAF (exon 15), TERT (promoter region), and GNAQ and GNA11 (exons 4 and 5) were performed using the Big Dye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA) in accordance with the manufacturer's instructions for the 3500XL Genetic Analyzer (Applied Biosystems). The primer sequences are listed in supplemental online Table 2. Mutations in BRAF exon 15 V600E were identified by the 7500 real‐time quantitative PCR system (Applied Biosystems) using the minor groove binder (MGB) probes. Primers and probes for the V600E assay were as follows: forward: 5’‐ATGAAGACCTCACAGTAAAAATAGG‐3’; reverse: 5’‐AGACAACTGTTCAAACTGATGGG‐3’; mutation anchor: FAM‐TCTAGCTACAGAGAAA‐MGB; wild anchor: HEX‐TCTAGCTACAGTGAAA‐MGB.
PD‐L1 Expression and Copy Number Detection
The determination of PD‐L1 expression was performed with the rabbit monoclonal anti‐PD‐L1 antibody (E1L3N; dilution 1:200; Cell Signaling Technology, Danvers, MA) using an Autostainer Plus (Dako; Agilent Technologies, Santa Clara, CA). PD‐L1 expressions in tumor cells and tumor‐infiltrating lymphocytes (TILs) were classified as positive if moderate‐to‐strong membrane staining was observed in >1% of the tumor cells and/or TILs. PD‐L1 gene copy number per cell was investigated by fluorescence in situ hybridization (FISH) using a PD‐L1/chromosome 9 centromere probe (LBP Medicine Science and Technology Co., Ltd, Guangzhou, China). FISH analysis was independently reviewed by two investigators who were blinded to the gene expression data (by X.Z. and X.‐H.Y.). The copy numbers were counted in 100 nonoverlapping tumor cell nuclei. As there is no consensus on a standard approach in the PD‐L1 FISH scoring system, for this study tumors with >5 PD‐L1 copies per cell were classified as PD‐L1 FISH positive (+) according to the Cappuzzo scoring system 40, 41, including PD‐L1 amplification, which was characterized by tumor cells with PD‐L1‐CEP9 ratio > 2.0 or > 10 copies per cell in >10% tumor cells. PD‐L1 positive expression and/or amplification were regarded as a PD‐L1+. PD‐L1 loss was characterized by a PD‐L1‐CEP9 ratio of <0.8. The FISH signals were assessed under a microscope (Olympus BX61; Olympus, Tokyo, Japan) equipped with a triple‐pass filter (DAPI/Green/Orange, Vysis). Images were acquired using the BioView Automated Imaging Analysis System (BioView Ltd, Rehovot, Israel).
Statistical Analysis
Differences in the distributions of baseline characteristics were investigated using the chi‐squared (χ2) or Fisher's test between subgroups. Overall survival (OS) and 95% confidence intervals (CIs) were calculated using the Kaplan‐Meier method and compared using the log‐rank test. A Cox proportional hazard model was initially built for a univariate analysis and then used to evaluate independent factors for each biological and clinical feature associated with survival. All statistical analyses were performed using the SPSS software version 19.0 for Windows (SPSS Inc., Chicago, IL), and statistical significance was defined as a probability level <.05.
Results
Patients’ Clinical Characteristics
The main clinicopathological and molecular features of the 36 vaginal melanomas are summarized in Table 1 and supplemental online Table 2. The median age of the patients was 48 years (range, 27–77). The predominant growth pattern observed was the nodular subtype (88.9%, 32/36), followed by the superficial spreading (5.6%, 2/36). Twenty‐eight cases (77.8%) showed an epithelioid morphology, and five (13.9%) displayed a spindle cell morphology. Pigmentation was observed in 33 vaginal melanomas (91.7%). The DOI of the tumors ranged from 0.4 to 62.5 mm (median, 10.0), and ulceration were observed in 52.8% of the cases (19/36). Mitotic activity was 0 to 80 per mm2 (median, 8 per mm2).
Table 1.
Clinicopathologic characteristics of the 36 primary vaginal melanomas
| Variable | Patients, n (%) |
|---|---|
| Total | 36 |
| Median age (range), yr | 48 (27–77) |
| Tumor phenotype | |
| Superficial spreading | 2 (5.6) |
| Nodular | 32 (88.9) |
| Unknown | 2 (5.6) |
| Ulceration | |
| Absent | 17 (47.2) |
| Present | 19 (52.8) |
| Cellularity | |
| Epitheloid | 28 (77.8) |
| Spindle cell | 5 (13.9) |
| Mixed | 3 (8.3) |
| Pigmentation | |
| Absent | 3 (8.3) |
| Present | 33 (91.7) |
| Mitotic activity, n/mm2 | |
| 0 | 4 (11.1) |
| 1–10 | 16 (44.4) |
| >10 | 16 |
| Breslow thickness, mm | 44.4 |
| Median (range) | 10 (0.4–62.5) |
| ≤1.0 | 1 (2.8) |
| 1.0–2.0 | 2 (5.6) |
| 2.0–4.0 | 3 (8.3) |
| 4.0–10.0 | 13 (36.1) |
| >10.0 | 14 (38.9) |
| Unknown | 3 (8.3) |
| Surgery approach | |
| WLE | 17 (47.2) |
| RE | 15 (41.7) |
| Biopsy | 4 (11.1) |
| Lymphadenectomy | |
| Yes | 26 (72.2) |
| No | 10 (27.8) |
| Lymph node metastasis | |
| Present | 10 (27.8) |
| Absent | 22 (61.1) |
| Unknown | 4 (11.1) |
| AJCC Stage | |
| I | 2 (5.6) |
| II | 22 (61.1) |
| III | 10 (27.8) |
| Unknown | 2 (5.6) |
Abbreviations: AJCC, American Joint Committee on Cancer; RE, Radical excision; WLE, wide local excision.
The Prevalence of Oncogenic Mutations
Direct sequencing was performed to identify the status of the gene mutations. NRAS mutations were found in 5 of the 36 (13.9%) primary melanomas, of which 4 had Q61R mutation, and one had Q61P mutation. No clinicopathological feature demonstrated any significant association with the NRAS mutation status (supplemental online Table 3). KIT sequence analysis was performed in 34 cases, of which a missense and V559D mutation in KIT exon 11 was detected in one patient (2.9%, 1/34). TERT C228T mutations were identified in two cases (5.6%), one of whom had a concurrent NRAS Q61R mutation. BRAF, GNAQ, and GNA11 mutations were not detected in any cases. Samples of the sequencing electropherograms of cases demonstrating NRAS, KIT, and TERT mutated genes are shown in supplemental online Figure 1.
PD‐L1 Expression and Copy Number Alterations
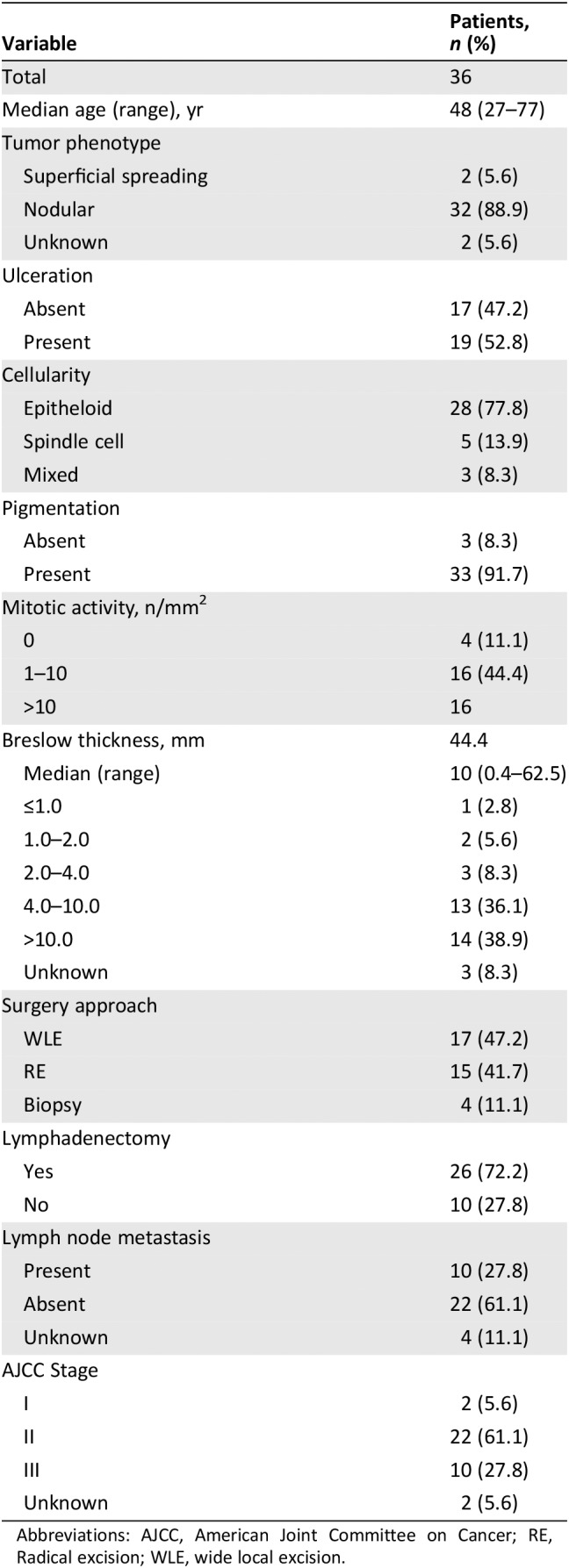
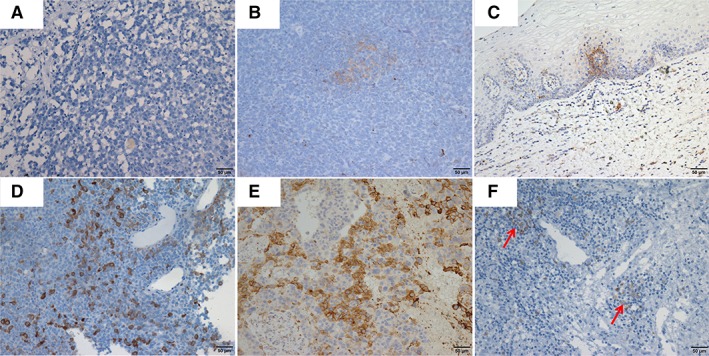
The representative images for IHC and FISH are illustrated in Figures 1 and 2, respectively. The IHC method for PD‐L1 expression demonstrated that 27.8% of patients (10/36) exhibited PD‐L1 focal staining, of these, the percentage of tumor cell staining was 3% in three cases, 5% in 3, 8% in one, 20% in one, and 30% in one, whereas PD‐L1 positive expression of TILs was observed in only one patient (Fig. 1; Table 2). PD‐L1 FISH+ was found in three patients, of whom two harbored PD‐L1 amplifications defined by a ratio of 2.23 and 2.67 (Fig. 2D and E; Table 2). By contrast, there was one patient with PD‐L1 loss (PD‐L1/CEP9 ratio = 0.74; Fig. 2F; Table 2). Interestingly, there were two cases (case number 28 [PD‐L1 copy numbers/ratio, 4.2/2.23] and 32 [PD‐L1 copy numbers/ratio, 6.0/1.33]) in which no PD‐L1 expression was detected but showed PD‐L1 positivity in FISH (Table 2). PD‐L1+ was found to occur more frequently in older patients (> 60 years; p = .008) but showed no significant association with other clinicopathological features (supplemental online Table 3).
Figure 1.
Photomicrographs showing immunohistochemistry staining of the programmed death‐ligand 1 (PD‐L1) expression of tumor cells and tumor‐infiltrating lymphocytes (TILs) in vaginal melanoma samples. The percentage of PD‐L1 expression in tumor cells was 0% (A), 3% (B and C), 20% (D), and 30% (E). PD‐L1‐positive staining in TILs was indicated by arrows (F).
Figure 2.
The representative images of the programmed death‐ligand 1 (PD‐L1) copy number changes detected by fluorescence in situ hybridization. PD‐L1 signals with less than five copies per cell were detected in patients (A and B). Six PD‐L1 signals per cell were identified in case number 32 (C); 4.2 copies but ratio equals to 2.23 were determined in case number 28 (D); 10.7 copies and ratio equals to 2.67 were identified in case number 19 (E), whereas PD‐L1 loss was found in case number 33 (F).
Table 2.
Clinical, pathological, and molecular features of the investigated 36 patients with primary vaginal melanomas
| Case no. | Age | Surgical approach | Tumor type | Ulceration | Cellularity | Pigmentation | Mitotic activity, n/mm2 | Depth of invasion, mm | AJCC Stage | Molecular findings | PD‐L1 expression | PD‐L1 copy numbers/ratio | Follow‐up time, mo | Follow‐up status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 36 | RE | NM | Present | E | Present | 8 | 8.2 | pT4bN0M0, IIC | NRAS p.Q61R | No staining | 2.1/1.06 | 14.0 | DOD |
| 2 | 72 | WLE | SSM | Absent | E | Present | 0 | 0.4 | pT1aN0M0, IA | None | TC, 3% | 2.1/1.13 | 32.2 | DOD |
| 3 | 47 | RE | NM | Absent | E | Present | 24 | 27.5 | pT4aN1M0, IIIC | NRAS p.Q61R | No staining | 2.0/1.03 | 8.9 | DOD |
| 4 | 69 | WLE | NM | Present | E | Present | 3 | 1.8 | pT2bN0M0, IIA | None | No staining | 2.3/1.23 | 17.2 | DOD |
| 5 | 67 | RE | NM | Present | E | Present | 7 | 3.5 | pT3bN0M0, IIB | None | No staining | 3.5/1.56 | 15.0 | DOD |
| 6 | 74 | WLE | NM | Present | S | Present | 0 | 15.0 | pT4bN0M0, IIC | NRAS p.Q61R | No staining | 2.0/1.03 | 42.7 | DOD |
| 7 | 40 | Biopsy | NA | Present | E | Present | 6 | 7.4 | cT4bN0M0, IIC | KIT p.V559D | No staining | 2.2/1.05 | Unknown | Unknown |
| 8 | 41 | RE | NM | Present | E | Absent | 1 | 35.0 | pT4bN0M0, IIC | None | TC, 3% | 2.0/1.06 | 8.2 | DOD |
| 9 | 42 | RE | NM | Present | S | Present | 4 | 14.3 | pT4bN1M0, IIIC | None | No staining | 2.0/1.04 | 40.3 | DOD |
| 10 | 46 | WLE | NM | Absent | E | Present | 2 | 9.1 | pT4aN3M0, IIIC | TERT C228T | No staining | 2.1/1.07 | 11.5 | DOD |
| 11 | 33 | Biopsy | NM | Absent | E | Present | 8 | Unknown | Unknown | None | No staining | 2.8/1.24 | 6.2 | DOD |
| 12 | 64 | WLE | NM | Present | E | Present | 0 | 7.9 | pT4bN0M0, IIC | None | No staining | 2.0/1.04 | 66.4 | DOD |
| 13 | 38 | RE | NM | Present | E | Present | 6 | 16.3 | pT4bN1M0, IIIC | NRAS p.Q61R TERT C228T | No staining | 3.0/1.20 | 6.5 | DOD |
| 14 | 60 | WLE | NM | Absent | E | Present | 13 | 10.7 | pT4aN0M0, IIB | None | No staining | 2.2/1.11 | 56.5 | DOD |
| 15 | 59 | WLE | NM | Present | E | Present | 8 | 17.8 | pT4bN1M0, IIIC | None | No staining | 2.0/1.05 | 7.5 | DOD |
| 16 | 43 | WLE | NM | Absent | S | Present | 14 | 7.0 | pT4aN0M0, IIB | None | No staining | 2.0/1.03 | 6.9 | DOD |
| 17 | 57 | WLE | NM | Absent | E | Absent | 0 | 4.0 | pT3aN0M0, IIA | None | No staining | 3.9/1.37 | 33.5 | DOD |
| 18 | 47 | RE | NM | Present | E | Present | 4 | 8.6 | pT4bN1M0, IIIC | None | No staining | 2.3/1.14 | 29.9 | DOD |
| 19 | 67 | WLE | NM | Absent | E | Present | 80 | 2.0 | pT2aN0M0, IB | None | TC, 8% | 10.7/2.67 | 14.5 | A/L |
| 20 | 65 | RE | NM | Absent | E | Present | 15 | 12.0 | pT4aN0M0, IIB | None | TIL staining | 2.0/1.03 | 19.6 | A/L |
| 21 | 68 | WLE | NM | Absent | E | Present | 3 | 9.0 | pT4aN0M0, IIB | None | TC, 5% | 2.2/1.05 | 15.0 | A/L |
| 22 | 34 | RE | NM | Absent | E | Present | 23 | 63.0 | pT4aN2M0, IIIC | None | TC, 5% | 2.0/1.04 | 13.9 | A/L |
| 23 | 44 | Biopsy | NM | Absent | E | Present | 5 | Unknown | Unknown | None | TC, 20% | 2.1/1.03 | 20.0 | A/L |
| 24 | 39 | RE | NM | Absent | E | Present | 7 | 10.0 | pT4aN0M0, IIB | None | No staining | 2.0/1.04 | 9.8 | A/L |
| 25 | 70 | WLE | NM | Present | S | Present | 29 | 13.0 | pT4bN0M0, IIC | None | TC, 30% | 3.2/1.23 | 7.8 | A/L |
| 26 | 46 | RE | NM | Absent | E | Present | 17 | 51.5 | pT4aN3M0, IIIC | None | No staining | 3.4/1.20 | 82.9 | A/L |
| 27 | 45 | RE | NM | Present | E | Present | 21 | 17.2 | pT4bN1M0, IIIC | None | TC, 5% | 2.0/1.02 | 12.9 | A/L |
| 28 | 73 | WLE | NM | Present | E + S | Present | 65 | 27.5 | pT4bN0M0, IIC | None | No staining | 4.2/2.23 | 5.4 | A/L |
| 29 | 52 | Biopsy | NA | Present | E | Present | 30 | > 4.0 | cT3bN0M0, IIB | None | No staining | 3.7/1.24 | 25.4 | A/L |
| 30 | 50 | RE | NM | Present | E | Present | 17 | 10.0 | pT4bN0M0, IIC | None | No staining | 2.2/1.09 | 23.6 | DOD |
| 31 | 48 | RE | NM | Present | E | Present | 16 | 3.0 | pT3bN0M0, IIB | None | No staining | 2.0/1.04 | 69.0 | A/L |
| 32 | 47 | WLE | NM | Absent | E | Present | 10 | 9.0 | pT4aN1M0, IIIC | None | No staining | 6.0/1.33 | 70.2 | DOD |
| 33 | 51 | WLE | SSM | Absent | E | Present | 12 | 5.0 | pT4aN0M0, IIB | NRAS p.Q61P | No staining | 2.1/0.74 | 15.7 | DOD |
| 34 | 27 | WLE | NM | Present | E + S | Present | 8 | 10.0 | pT4bN0M0, IIC | None | No staining | 2.0/1.04 | 43.3 | DOD |
| 35 | 77 | WLE | NM | Present | E + S | Present | 12 | 15.0 | pT4bN0M0, IIC | None | TC, 3% | 3.8/1.22 | 9.5 | A/L |
| 36 | 48 | RE | NM | Absent | S | Absent | 20 | 5.5 | pT4aN0M0, IIB | None | No staining | 2.0/1.06 | 8.4 | A/L |
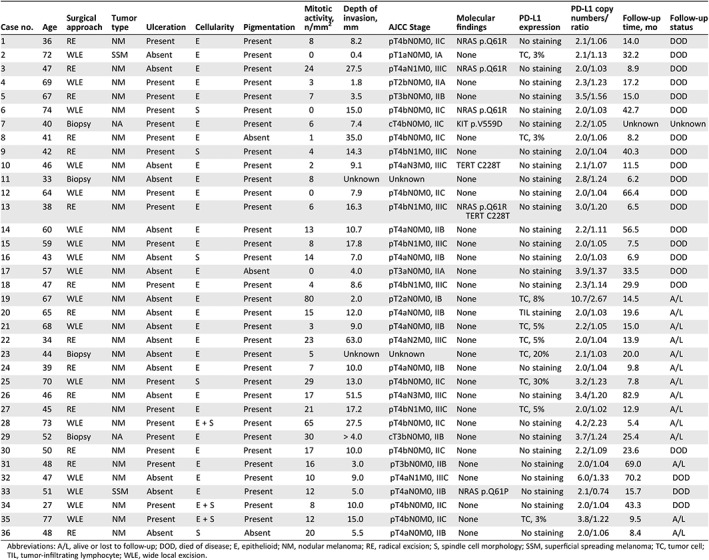
Abbreviations: A/L, alive or lost to follow‐up; DOD, died of disease; E, epithelioid; NM, nodular melanoma; RE, radical excision; S, spindle cell morphology; SSM, superficial spreading melanoma; TC, tumor cell; TIL, tumor‐infiltrating lymphocyte; WLE, wide local excision.
Patients’ Survival
Follow‐up information was available for 35 patients, among whom 27 (77.1%) developed recurrences and metastases, and 21 (60.0%) died. Furthermore, Kaplan‐Meier survival analyses showed that patients with mutated NRAS had a worse OS compared with those with a wild‐type NRAS (33.5 vs. 14.0 months; HR, 3.09; 95% CI, 1.08–8.83; p = .035; Fig. 3A), whereas no statistical significance in OS was observed for patients with PD‐L1 expression and/or PD‐L1 amplification compared with those without expression/amplification (Fig. 3B). Multivariate analyses demonstrated that >10 mitoses per mm2 (HR, 2.96; 95% CI, 1.03–8.51; p = .043) was an independent prognostic factor in patients with primary vaginal melanomas (Table 3).
Figure 3.
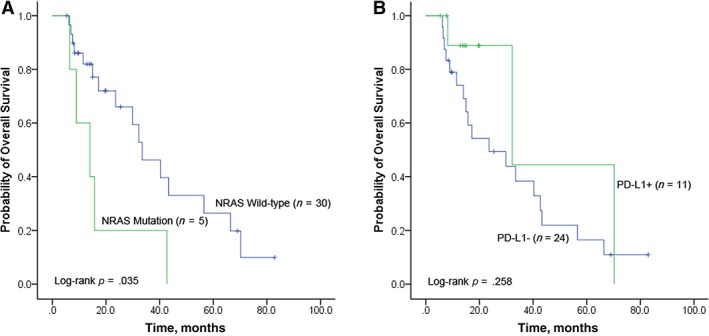
Kaplan‐Meier curves for overall survival (OS) in the 36 investigated patients with primary vaginal melanoma. (A): Patients with wild‐type NRAS had a favorable OS than those with mutated NRAS (p = .035). (B): No statistical significance for OS between patients with/without programmed death‐ligand 1 (PD‐L1) positive staining and/or amplifications was found.
Table 3.
Univariate and multivariate Cox Regression analyses for overall survival in primary vaginal melanoma
| Variables | Univariate HR (95% CI) | p valuea | Multivariate HR (95% CI) | p valuea |
|---|---|---|---|---|
| Age, years (≤60 vs. >60) | 1.32 (0.51–3.45) | .571 | ||
| Tumor type (NM vs. SSM) | 1.84 (0.41–8.27) | .426 | ||
| Ulceration (present vs. absent) | 0.73 (0.30–1.79) | .488 | ||
| Cellularity (epithelioid vs. spindle) | 0.97 (0.32–2.99) | .956 | ||
| Pigmentation (present vs. absent) | 1.86 (0.42–8.33) | .417 | ||
| DOI (≤4 mm vs. >4 mm) | 0.87 (0.29–2.61) | .796 | ||
| Mitotic activity (≥10 vs. <10) | 3.24 (1.15–9.19) | .027 | 2.96 (1.03–8.51) | .043 |
| LN metastasis (absent vs. present) | 0.99 (0.37–2.62) | .975 | ||
| Clinical Stage (I vs. II + III) | 1.02 (1.32–7.87) | .985 | ||
| NRAS status (mut vs. wt) | 3.09 (1.08–8.83) | .035 | 2.58 (0.89–7.43) | .080 |
| PD‐L1 status (PD‐L1+ vs. PD‐L1−)b | 2.05 (0.59–7.07) | .258 |
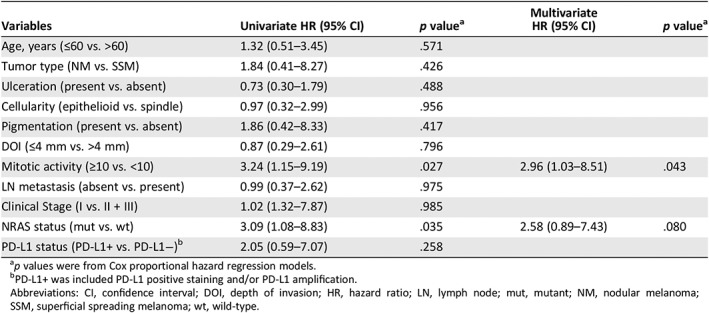
p values were from Cox proportional hazard regression models.
PD‐L1+ was included PD‐L1 positive staining and/or PD‐L1 amplification.
Abbreviations: CI, confidence interval; DOI, depth of invasion; HR, hazard ratio; LN, lymph node; mut, mutant; NM, nodular melanoma; SSM, superficial spreading melanoma; wt, wild‐type.
Two cases were prescribed immune checkpoint inhibitors as subsequent treatment after wide local excision or chemoradiotherapy. The first one, case number 21, showed a 5% positive staining for PD‐L1 and was treated by nivolumab as adjuvant therapy (1 mg/kg every 3 weeks). The patient relapsed, identified by computed tomography (CT) imaging, after 7 months but still continued the nivolumab therapy, then changed to pembrolizumab after one cycle, and at last follow‐up, Dec 24, 2018, the tumor was observed to have shrunken (13 × 16 mm to 13 × 10 mm). The second case, case number 29, did not show any PD‐L1 expression. Upon completion of chemoradiotherapy, the patient was diagnosed with liver metastasis and underwent 11 cycles with pembrolizumab (10 mg/kg every 2 weeks). During the treatment, brain metastasis was identified on CT peri‐treatment follow‐up examination and the patient was given concurrent radiotherapy. Fourteen months later, the tumor was observed by abdominal ultrasound to have metastasized to the liver, and the patient was offered ablation therapy. During the course of the treatment, pembrolizumab was continuously given, and at last follow‐up, Oct 18, 2018, the patient was in good condition, Eastern Cooperative Oncology Group (ECOG) performance status 1.
Discussion
Little information is known about the molecular characteristics in primary vaginal melanoma, although it has been demonstrated that mucosal melanoma is genetically distinct from cutaneous melanoma and more commonly exhibits KIT and NRAS mutations. We reviewed 36 cases of vaginal melanomas and found a relatively low frequency of genetic mutations. NRAS was the most mutated gene in primary vaginal melanomas and was associated with worse OS. PD‐L1 expression was observed commonly. We also reported the patients with or without PD‐L1 expression may benefit from immune checkpoint inhibitors. These findings offer insights therapeutic targets for these patients.
In contrast to existing literature, NRAS mutations were frequently found in melanomas of the skin with CSD and had mutation rates of up to 24% 42. The frequency of NRAS mutations varied among mucosal melanomas, with a wide range of 0%–43% mutation rates reported in previous studies 10, 43, 44, 45, 46. It was found that NRAS mutations were present in 37.5% (6/16) of esophageal mucosal melanomas 47 and 7.1%–30% of sinonasal melanomas 48, 49, 50, and none were detected until now in oral mucosal melanomas 50, 51. In melanomas of the female urogenital tract, there were two studies showing the different mutation rates of 13.3% (2/15) 43 and 21.4% (3/14) 44. In our series, NRAS mutations were detected in 13.9% of vaginal melanomas. Previously, NRAS mutations have been observed to be associated with some features predictive of aggressive behavior in cutaneous melanomas, such as the Clark level of invasion, Breslow thickness, ulceration rate, and adverse outcome 15, 16. However, the prognostic value of NRAS status in melanoma is still a matter of intense debate. Several studies have been carried out to examine the effect of NRAS mutations on clinical outcomes and none of them found any impact on OS 52, 53, 54. In contrast, Devitt et al. reported that NRAS mutations were an adverse prognostic factor in multivariate analysis in a prospective cohort of 249 patients with melanoma 55. Besides, most of the observations have been conducted in white populations, with a scarce report from other geographic areas like Asia. In the present study, it was found that patients with mutated NRAS had inferior OS compared with those with wild‐type NRAS, although NRAS mutation status but had no significant correlation with any of the investigated clinicopathological features.
In previous studies, Beadling et al. 56 found KIT mutations in 15.6% of mucosal melanomas, and Curtin et al. 19 found KIT mutations and/or an increase in the copy number of KIT in 39.0% of mucosal melanomas. In contrast to our present study, only one patient (2.9%) was found to harbor the KIT V559D mutation, which was located in the juxtamembrane domain of the KIT receptor. Our finding is in line with previous studies that have also reported a low frequency (0%–8.3%) of KIT mutations, with only one case reported in vaginal melanomas (supplemental online Table 4). Patients with melanoma with this type of mutation were reported to benefit from imatinib 57, whereas a recent ECOG phase II trial investigating the use of dasatinib in mucosal melanomas with KIT alterations revealed low response rate 58, demonstrating that molecularly targeted therapy may not always be effective. Advancement in the therapeutics of these patients are difficult considering the rarity of mucosal melanomas and that only few of such patients do present with KIT mutations, thereby hindering the launch of large clinical trials to verify the effectiveness of KIT inhibitors.
No patients were found with mutations in BRAF exon 15 in the present study, which was similar with other published data on vaginal melanoma 10, 43, 44, 46. BRAF V600E mutation is the most common mutation in melanomas, with reports of up to a 40% prevalence in cutaneous melanoma 14, 15. However, in mucosal melanoma, BRAF mutations occur at a lower frequency (3.0%–15.5% in unspecialized mucosal melanoma) 15, 19. This is similar for vaginal melanoma, in which only four cases of BRAF mutations have been reported to date; one case was found in Aulmann et al.'s study 43, and three cases were in one study conducted by Hou et al. 45. Mutation in GNAQ/GNA11 exhibits tissue specificity in melanomas. No mutations were observed in the present study (vaginal melanoma) and in sinonasal and oral mucosal melanomas 50, 51, whereas it was demonstrated that the GNAQ/GNA11 active mutation is a major contributor in the development of uveal melanoma 21, 22
TERT promoter mutations were recently identified at high frequencies in cutaneous malignant melanoma tumor samples 24, whereas few have shown a low frequency of TERT promoter mutations in mucosal melanomas. Three studies have reported a relatively low frequencies of these mutations in unspecified location of mucosal melanomas: 23% (6/26) 25, 13.2% (7/53) 59, and 12.5% (1/8) 26. In specific location of mucosal melanomas, namely sinonasal malignant melanomas, the frequencies of TERT mutations were separately identified as 8% (4/49) 60 and 11.5% (3/28) 50. To our knowledge, this present study is the first investigate the TERT promoter mutations in vaginal melanomas. Two patients (7.7%) with primary tumors were diagnosed as stage IIIC vaginal melanomas with TERT promoter mutations in C228T and had an overall survival of less than 12 months. Moreover, TERT promoter mutations have been reported to be associated with older patients, increased Breslow thickness, and worse prognosis 25, 26. By contrast, an investigation from Asia demonstrated that the TERT promoter mutations were not correlated with OS 61. The difference in frequencies and relationship with clinicopathological features reveal that the TERT promoter mutations may vary depending on the melanoma subtypes and locations.
Although vulvar melanoma arises on hairy and glabrous skin of the vulva, it was described as a mucosal melanoma because of its continuity with the vaginal mucosa and its low‐sunlight exposure location 62. Several studies have characterized the molecular events of melanomas in both the vulva and vagina. Hou et al. 45 assessed all the reported cases of vulvar and vaginal melanoma with molecular detection and concluded that these two types of melanomas had distinct molecular signatures. The genes most commonly mutated in vulvar melanomas were KIT (26.5%, 9/34) and BRAF (27%, 9/33), whereas NRAS mutations were more prevalent than KIT and BRAF mutations in vaginal melanomas 45. Although the varying mutation frequencies might be related to the limited number of cases analyzed and differences in methodology in all of those studies, the different frequencies of molecular alterations suggest that the development of vulvar and vaginal melanomas involves different tumorigenesis pathways. This finding is essential for patients with vaginal melanomas who harbor frequent NRAS mutations because they might benefit from MEK inhibition 30.
Accordingly, the potential clinical implications for testing molecular alterations and targeted treatments were further explored in this study. The regulation of immune checkpoint has been recently investigated in a variety of malignancies. In previous clinical trials, a number of monoclonal antibodies against PD‐1 and PD‐L1 with antitumor activities have been observed in some of patients with cutaneous melanoma 63, 64. A phase Ib KEYNOTE‐001 study has reported that PD‐L1 expression, assessed by IHC assay (clone, 22C3), is a potential predictive marker for anti‐PD‐L1 activity in patients with advanced melanoma who were treated with pembrolizumab 65. In the present study, we confirmed that 27.8% of the patients with vaginal melanoma were considered as having PD‐L1‐positive staining (E1L3N). From the medical records, one patient with PD‐L1‐positive staining received nivolumab treatment, whereas another patient without PD‐L1‐positive staining received pembrolizumab. Both of them were still alive with stable tumor response until the end of the follow‐up period. Because of the low incidence of mucosal melanoma, only limited data have been published regarding the efficacy of immune checkpoint inhibitors in this disease subtype. Previous reports have suggested that the response rates to ipilimumab treatment in patients with advanced mucosal melanoma are much lower than in patients with cutaneous melanoma 66, 67. However, there are few investigations on the response of pembrolizumab and nivolumab in vaginal melanoma. Because of the small number of patients, it may be difficult to interpret the data. Nevertheless, this observation could act as a hint that patients with vaginal melanoma with PD‐L1 expression could benefit from immune checkpoint inhibitors even for those without PD‐L1 expression, which is in line with the report of a previous study 65.
Despite multiple effective treatment options for cutaneous melanoma, data on the treatment of vagina mucosa melanomas are limited, especially from Asia. Therefore, patients with vaginal melanomas should be encouraged to perform more comprehensive analysis, including that of NRAS and KIT, and PD‐L1 expression/copy number at the time of initial diagnosis to search for an effective target and participate in clinical trials involving treatments with novel targeted therapies based on molecular aberrations. Two patients had no PD‐L1 expression but harbored PD‐L1 FISH+ in our study. To our knowledge, PD‐L1 FISH might be a good complement even in the absence of PD‐L1 expression, suggesting that more people could benefit from the immune checkpoint inhibitors. This is in agreement with a recent study which showed that PD‐L1 amplified solid tumors had beneficial objective responses after administration of immune checkpoint inhibitors 68. Further investigation is required to determine whether PD‐L1 expression in TILs and/or tumor cells predict response to immunotherapy in patients with vaginal mucosal melanoma. PD‐L1 amplification did not always correlate with PD‐L1 expression by IHC analysis. Additional large‐scale, prospective studies of PD‐L1‐amplified cancers are warranted to confirm the responses to immune‐checkpoint blockade prescribed, even in the absence of PD‐L1 expression. Therefore, a coordinated, collaborative effort is required to collect more samples and further progress is to be made in understanding the pathogenesis and offering optimal treatment.
There were several limitations in our study. Vaginal mucosal melanomas are rare, with few existing large retrospective studies. The sample size was small and may have limited the univariate and multivariate analyses for predicting the correlation between clinical variables and gene status with oncogenic outcome. The definitive prognostic factors are yet to be fully determined using larger cohorts of patients.
Conclusion
We provided highlights of the molecular insights within the spectrum of vaginal melanomas. In this cohort of patients with primary vaginal melanoma, we observed that NRAS mutations and PD‐L1 expression were most prevalent, whereas the detection rate of KIT and TERT mutations was low. Patients with NRAS mutations had a poorer survival outcome as compared with those with wild‐type NRAS. No significant difference in OS was observed between those with and without PD‐L1 expression and amplification. The only clinicopathological feature identified as an independent factor for survival was mitotic activity.
Author Contributions
Conception/design: Hai‐Yun Wang, Fang Wang
Provision of study material or patients: Xiao Zhang, Yan‐Fen Feng
Collection and/or assembly of data: Xiao‐Yan Wu, Xin‐Hua Yang, Ya‐Kang Long
Data analysis and interpretation: Hai‐Yun Wang, Yan‐Fen Feng, Fang Wang
Manuscript writing: Hai‐Yun Wang, Fang Wang
Final approval of manuscript: Hai‐Yun Wang, Xiao‐Yan Wu, Xiao Zhang, Xin‐Hua Yang, Ya‐Kang Long, Yan‐Fen Feng, Fang Wang
Disclosures
The authors indicated no financial relationships.
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Supplementary Figure 1
Supplementary Tables
Acknowledgments
We would like to thank Dr. Seeruttun Sharvesh Raj for the professional English language revision of this manuscript and thank the patients who participated in our study.
Ethic approval was given by the Ethics Committee of the Sun Yat‐sen University Cancer Center (No.B2016‐069‐01). Informed consent was obtained from all individual participants included in this study. All procedures of this study involving human participants were performed in accordance with the ethical standards of the Sun Yat‐sen University Cancer Center and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
The raw data in this paper has been successfully uploaded onto the Research Data Deposit with RDD identifier number: RDDA2019001043.
This study was supported by the Guangdong Natural Science Foundation (No. 2017A030310192), Fundamental Research Funds for the Central Universities (No. 17ykpy84), National Natural Science Foundation of China (No. 31360258, 81602468), and Natural Science Foundation of Guangdong Province (No. 2015A030313047).
Disclosures of potential conflicts of interest may be found at the end of this article.
Contributor Information
Yan‐Fen Feng, Email: fengyf@sysucc.org.cn.
Fang Wang, Email: wangfang@sysucc.org.cn.
References
- 1. Erdmann F, Lortet‐Tieulent J, Schüz J et al. International trends in the incidence of malignant melanoma 1953‐2008–are recent generations at higher or lower risk? Int J Cancer 2013;132:385–400. [DOI] [PubMed] [Google Scholar]
- 2. Chang AE, Karnell LH, Menck HR. The national cancer data base report on cutaneous and noncutaneous melanoma: A summary of 84,836 cases from the past decade. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer 1998;83:1664–1678. [DOI] [PubMed] [Google Scholar]
- 3. Frumovitz M, Etchepareborda M, Sun CC et al. Primary malignant melanoma of the vagina. Obstet Gynecol 2010;116:1358–1365. [DOI] [PubMed] [Google Scholar]
- 4. Cobellis L, Calabrese E, Stefanon B et al. Malignant melanoma of the vagina. A report of 15 cases. Eur J Gynaecol Oncol 2000;21:295–297. [PubMed] [Google Scholar]
- 5. Reid GC, Schmidt RW, Roberts JA et al. Primary melanoma of the vagina: A clinicopathologic analysis. Obstet Gynecol 1989;74:190–199. [PubMed] [Google Scholar]
- 6. Xia L, Han D, Yang W et al. Primary malignant melanoma of the vagina: A retrospective clinicopathologic study of 44 cases. Int J Gynecol Cancer 2014;24:149–155. [DOI] [PubMed] [Google Scholar]
- 7. Huang Q, Huang H, Wan T et al. Clinical outcome of 31 patients with primary malignant melanoma of the vagina. J Gynecol Oncol 2013;24:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barth RF, Mi P, Yang W. Boron delivery agents for neutron capture therapy of cancer. Cancer Commun (Lond) 2018;38:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Simon A, Kourie HR, Kerger J. Is there still a role for cytotoxic chemotherapy after targeted therapy and immunotherapy in metastatic melanoma? A case report and literature review. Chin J Cancer 2017;36:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Omholt K, Grafstrom E, Kanter‐Lewensohn L et al. KIT pathway alterations in mucosal melanomas of the vulva and other sites. Clin Cancer Res 2011;17:3933–3942. [DOI] [PubMed] [Google Scholar]
- 11. Schoenewolf NL, Urosevic‐Maiwald M, Dummer R. Tumour heterogeneity of mucosal melanomas during treatment with imatinib. Br J Dermatol 2011;165:419–424. [DOI] [PubMed] [Google Scholar]
- 12. Curtin JA, Fridlyand J, Kageshita T et al. Distinct sets of genetic alterations in melanoma. N Engl J Med 2005;353:2135–2147. [DOI] [PubMed] [Google Scholar]
- 13. Edwards RH, Ward MR, Wu H et al. Absence of BRAF mutations in UV‐protected mucosal melanomas. J Med Genet 2004;41:270–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davies H, Bignell GR, Cox C et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–954. [DOI] [PubMed] [Google Scholar]
- 15. Si L, Kong Y, Xu X et al. Prevalence of BRAF V600E mutation in chinese melanoma patients: Large scale analysis of BRAF and NRAS mutations in a 432‐case cohort. Eur J Cancer 2012;48:94–100. [DOI] [PubMed] [Google Scholar]
- 16. Jakob JA, Bassett RL, Jr , Ng CS et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 2012;118:4014–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thomas NE, Edmiston SN, Alexander A et al. Association between NRAS and BRAF mutational status and melanoma‐specific survival among patients with higher‐risk primary melanoma. JAMA Oncol 2015;1:359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yaman B, Akalin T, Kandiloğlu G. Clinicopathological characteristics and mutation profiling in primary cutaneous melanoma. Am J Dermatopathol 2015;37:389–397. [DOI] [PubMed] [Google Scholar]
- 19. Curtin JA, Busam K, Pinkel D et al. Somatic activation of kit in distinct subtypes of melanoma. J Clin Oncol 2006;24:4340–4346. [DOI] [PubMed] [Google Scholar]
- 20. Van Raamsdonk CD, Bezrookove V, Green G et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009;457:599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Raamsdonk CD, Griewank KG, Crosby MB et al. Mutations in GNA11 in uveal melanoma. New Engl J Med 2010;363:2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lamba S, Felicioni L, Buttitta F et al. Mutational profile of GNAQQ209 in human tumors. PloS One 2009;4:e6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen X, Wu Q, Tan L et al. Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene 2014;33:4724–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang FW, Hodis E, Xu MJ et al. Highly recurrent TERT promoter mutations in human melanoma. Science 2013;339:957–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Griewank KG, Murali R, Puig‐Butille JA et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst 2014;106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roh MR, Park KH, Chung KY et al. Telomerase reverse transcriptase (TERT) promoter mutations in Korean melanoma patients. Am J Cancer Res 2017;7:134–138. [PMC free article] [PubMed] [Google Scholar]
- 27. Macerola E, Loggini B, Giannini R et al. Coexistence of TERT promoter and BRAF mutations in cutaneous melanoma is associated with more clinicopathological features of aggressiveness. Virchows Arch 2015;467:177–184. [DOI] [PubMed] [Google Scholar]
- 28. Long GV, Stroyakovskiy D, Gogas H et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF‐mutant melanoma: A multicentre, double‐blind, phase 3 randomised controlled trial. Lancet 2015;386:444–451. [DOI] [PubMed] [Google Scholar]
- 29. Kelleher FC, McArthur GA. Targeting NRAS in melanoma. Cancer J 2012;18:132–136. [DOI] [PubMed] [Google Scholar]
- 30. Ascierto PA, Schadendorf D, Berking C et al. MEK162 for patients with advanced melanoma harbouring NRAS or val600 BRAF mutations: A non‐randomised, open‐label phase 2 study. Lancet Oncol 2013;14:249–256. [DOI] [PubMed] [Google Scholar]
- 31. Fedorenko IV, Gibney GT, Smalley KS. NRAS mutant melanoma: Biological behavior and future strategies for therapeutic management. Oncogene 2013;32:3009–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hodi FS, Corless CL, Giobbie‐Hurder A et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun‐damaged skin. J Clin Oncol 2013;31:3182–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brahmer JR, Tykodi SS, Chow LQ et al. Safety and activity of anti‐PD‐L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Topalian SL, Hodi FS, Brahmer JR et al. Safety, activity, and immune correlates of anti‐PD‐1 antibody in cancer. N Engl J Med 2012;366:2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zou W, Wolchok JD, Chen L. Pd‐L1 (B7‐H1) and PD‐1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med 2016;8:328rv324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Day D, Hansen AR. Immune‐related adverse events associated with immune checkpoint inhibitors. BioDrugs 2016;30:571–584. [DOI] [PubMed] [Google Scholar]
- 37. Hu LY, Xu XL, Rao HL et al. Expression and clinical value of programmed cell death‐ligand 1 (PD‐L1) in diffuse large b cell lymphoma: A retrospective study. Chin J Cancer 2017;36:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Herbst RS, Soria JC, Kowanetz M et al. Predictive correlates of response to the anti‐PD‐L1 antibody MPDL3280A in cancer patients. Nature 2014;515:563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heintz AP, Odicino F, Maisonneuve P et al. Carcinoma of the ovary. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet 2006;95(suppl 1):S161–192. [DOI] [PubMed] [Google Scholar]
- 40. Tong JH, Yeung SF, Chan AW et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non‐small cell lung carcinoma with poor prognosis. Clin Cancer Res 2016;22:3048–3056. [DOI] [PubMed] [Google Scholar]
- 41. Go H, Jeon YK, Park HJ et al. High MET gene copy number leads to shorter survival in patients with non‐small cell lung cancer. J Thorac Oncol 2010;5:305–313. [DOI] [PubMed] [Google Scholar]
- 42. Wang HY, Sun BY, Zhu ZH et al. Eight‐signature classifier for prediction of nasopharyngeal carcinoma survival. J Clin Oncol 2011;29:4516–4525. [DOI] [PubMed] [Google Scholar]
- 43. Aulmann S, Sinn HP, Penzel R et al. Comparison of molecular abnormalities in vulvar and vaginal melanomas. Mod Pathol 2014;27:1386–1393. [DOI] [PubMed] [Google Scholar]
- 44. van Engen‐van Grunsven AC , Kusters‐Vandevelde HV et al. NRAS mutations are more prevalent than KIT mutations in melanoma of the female urogenital tract–A study of 24 cases from the Netherlands. Gynecol Oncol 2014;134:10–14. [DOI] [PubMed] [Google Scholar]
- 45. Hou JY, Baptiste C, Hombalegowda RB et al. Vulvar and vaginal melanoma: A unique subclass of mucosal melanoma based on a comprehensive molecular analysis of 51 cases compared with 2253 cases of nongynecologic melanoma. Cancer 2017;123:1333–1344. [DOI] [PubMed] [Google Scholar]
- 46. Rouzbahman M, Kamel‐Reid S, Al Habeeb A et al. Malignant melanoma of vulva and vagina: A histomorphological review and mutation analysis–A single‐center study. J Low Genit Tract Dis 2015;19:350–353. [DOI] [PubMed] [Google Scholar]
- 47. Sekine S, Nakanishi Y, Ogawa R et al. Esophageal melanomas harbor frequent NRAS mutations unlike melanomas of other mucosal sites. Virchows Arch 2009;454:513–517. [DOI] [PubMed] [Google Scholar]
- 48. Amit M, Tam S, Abdelmeguid AS et al. Mutation status among patients with sinonasal mucosal melanoma and its impact on survival. Br J Cancer 2017;116:1564–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zebary A, Jangard M, Omholt K et al. KIT, NRAS and BRAF mutations in sinonasal mucosal melanoma: A study of 56 cases. Br J Cancer 2013;109:559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ozturk Sari S, Yilmaz I, Taskin OC et al. BRAF, NRAS, KIT, TERT, GNAQ/GNA11 mutation profile analysis of head and neck mucosal melanomas: A study of 42 cases. Pathology 2017;49:55–61. [DOI] [PubMed] [Google Scholar]
- 51. Lyu J, Wu Y, Li C et al. Mutation scanning of BRAF, NRAS, KIT, and GNAQ/GNA11 in oral mucosal melanoma: A study of 57 cases. J Oral Pathol Med 2016;45:295–301. [DOI] [PubMed] [Google Scholar]
- 52. Akslen LA, Puntervoll H, Bachmann IM et al. Mutation analysis of the EGFR‐NRAS‐BRAF pathway in melanomas from black Africans and other subgroups of cutaneous melanoma. Melanoma Res 2008;18:29–35. [DOI] [PubMed] [Google Scholar]
- 53. Edlundh‐Rose E, Egyhazi S, Omholt K et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: A study based on mutation screening by pyrosequencing. Melanoma Res 2006;16:471–478. [DOI] [PubMed] [Google Scholar]
- 54. Ellerhorst JA, Greene VR, Ekmekcioglu S et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin Cancer Res 2011;17:229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dutton‐Regester K, Kakavand H, Aoude LG et al. Melanomas of unknown primary have a mutation profile consistent with cutaneous sun‐exposed melanoma. Pigment Cell Melanoma Res 2013;26:852– 860. [DOI] [PubMed] [Google Scholar]
- 56. Beadling C, Jacobson‐Dunlop E, Hodi FS et al. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res 2008;14:6821–6828. [DOI] [PubMed] [Google Scholar]
- 57. Heinrich MC, Corless CL, Demetri GD et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 2003;21:4342–4349. [DOI] [PubMed] [Google Scholar]
- 58. Kalinsky K, Lee S, Rubin KM et al. A phase 2 trial of dasatinib in patients with locally advanced or stage IV mucosal, acral, or vulvovaginal melanoma: A trial of the ECOG‐ACRIN cancer research group (E2607). Cancer 2017;123:2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Egberts F, Kruger S, Behrens HM et al. Melanomas of unknown primary frequently harbor TERT‐promoter mutations. Melanoma Res 2014;24:131–136. [DOI] [PubMed] [Google Scholar]
- 60. Jangard M, Zebary A, Ragnarsson‐Olding B et al. TERT promoter mutations in sinonasal malignant melanoma: A study of 49 cases. Melanoma Res 2015;25:185–188. [DOI] [PubMed] [Google Scholar]
- 61. Bai X, Kong Y, Chi Z et al. MAPK pathway and TERT promoter gene mutation pattern and its prognostic value in melanoma patients: A retrospective study of 2,793 cases. Clin Cancer Res 2017;23:6120–6127. [DOI] [PubMed] [Google Scholar]
- 62. Hiratsuka J, Kamitani N, Tanaka R et al. Boron neutron capture therapy for vulvar melanoma and genital extramammary Paget's disease with curative responses. Cancer Commun (Lond) 2018;38:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hamid O, Robert C, Daud A et al. Safety and tumor responses with lambrolizumab (anti‐PD‐1) in melanoma. N Engl J Med 2013;369:134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ribas A, Hamid O, Daud A et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA 2016;315:1600–1609. [DOI] [PubMed] [Google Scholar]
- 65. Daud AI, Wolchok JD, Robert C et al. Programmed death‐ligand 1 expression and response to the anti‐programmed death 1 antibody pembrolizumab in melanoma. J Clin Oncol 2016;34:4102–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Alexander M, Mellor JD, McArthur G et al. Ipilimumab in pretreated patients with unresectable or metastatic cutaneous, uveal and mucosal melanoma. Med J Aust 2014;201:49–53. [DOI] [PubMed] [Google Scholar]
- 67. Del Vecchio M, Di Guardo L, Ascierto PA et al. Efficacy and safety of ipilimumab 3mg/kg in patients with pretreated, metastatic, mucosal melanoma. Eur J Cancer 2014;50:121–127. [DOI] [PubMed] [Google Scholar]
- 68. Goodman AM, Piccioni D, Kato S et al. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA Oncol 2018;4:1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Supplementary Figure 1
Supplementary Tables


