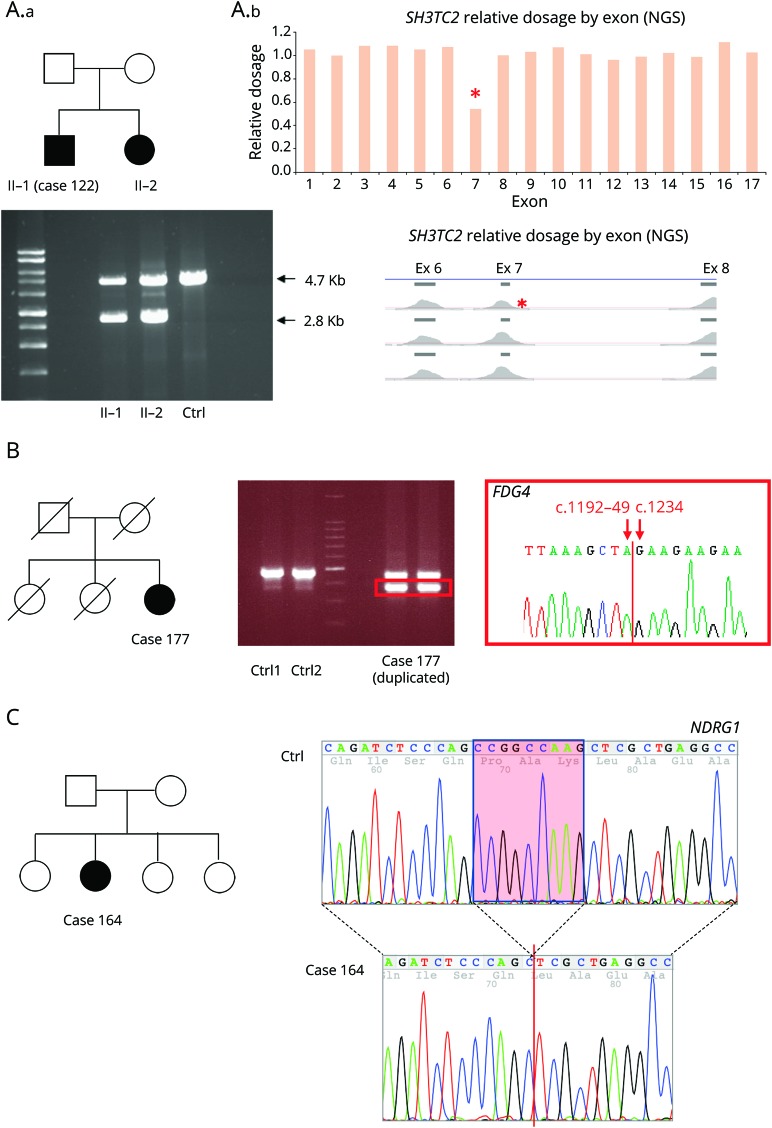
Figure 2. Representative examples of ancillary testing to next-generation sequencing (NGS) performed in selected cases.
(A) Case 122 presented with early onset of demyelinating neuropathy associated with scoliosis and cranial nerve involvement. He had a sister with a similar condition. NGS for genes associated with Charcot-Marie-Tooth disease type 1 (CMT1) and intermediate was performed and identified a single c.386-2A>C mutation in SH3TC2. Relative read-depth analysis of NGS was performed (A.b) looking for copy number variant in SH3TC2 and identified a deletion of exon 7 (indicated by a red * on the read depth plots), which was confirmed by long PCR in both siblings (A.a), in compound heterozygous state with the c.386-2A>C. (B) Patient 139 was diagnosed in the first decade of life with CMT1. A targeted NGS panel was performed at age 72, which identified 2 variants in FDG4 1304_1305delinsAA p.(Arg435Gln) and FDG4:c.1192-48_1233del. Long-range PCR was performed followed by Sanger sequencing of the gel band-extracted PCR product (red square box) identifying the breakpoints of 90–base pair FGD4 deletion. (C) Patient 164 presented with early-onset CMT1. NGS targeted panel for CMT1 genes was performed at age 35 and identified a homozygous 892-1 G>T variant in NDRG1, bearing potential to disrupt splicing of the flanking exons. RNA was extracted from peripheral blood and retrotranscribed into cDNA showing that the splicing mutation leads to a 9–base pair deletion of NDRG1 transcript (c.892_900delCCGGCCAAG) resulting in an in-frame deletion of 3 amino acids (red box). As opposed to typical CMT4D cases due to stop mutations in NDRG1, patient 164 presented a relatively mild neuropathy without clinical evidence of hearing loss, suggesting that the splicing mutation leading to in-frame deletion of 3 amino acidic residues may not abolish NDRG1 function.