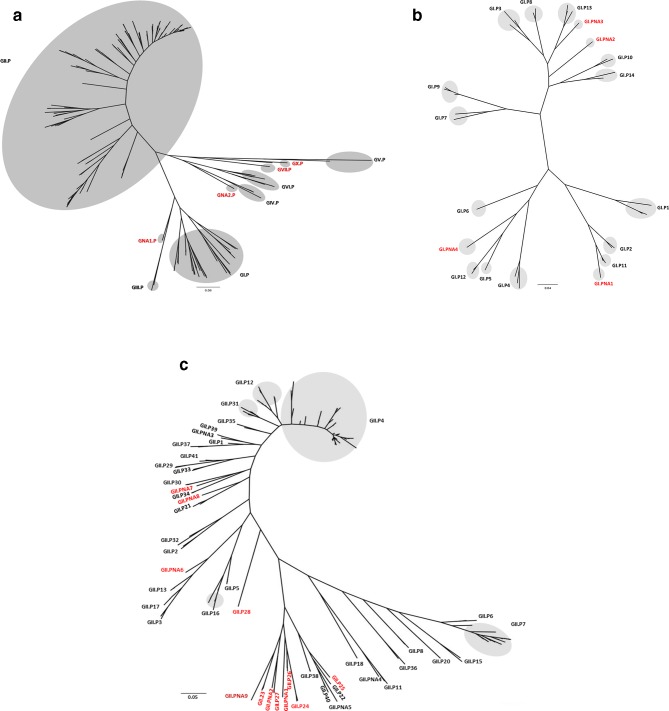
Fig. 3.
Phylogenetic trees of partial (762 nucleotides at 3′-end of ORF1) RdRp sequences of norovirus genogroups (a), GI P-types (b) and GII P-types (c). Tentative P-groups and P-types with only a single sequence or multiple non-identical sequences from a single geographic location are referred to as non-assigned (NA). Phylogenetic analysis was carried out using maximum likelihood (ML) with PhyML. The resulting trees were plotted and edited in FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Newly identified genogroups and genotypes are labelled in red. In Fig 3c, only GII P-types with highly divergent clusters are shown with a gray circle for clarity.