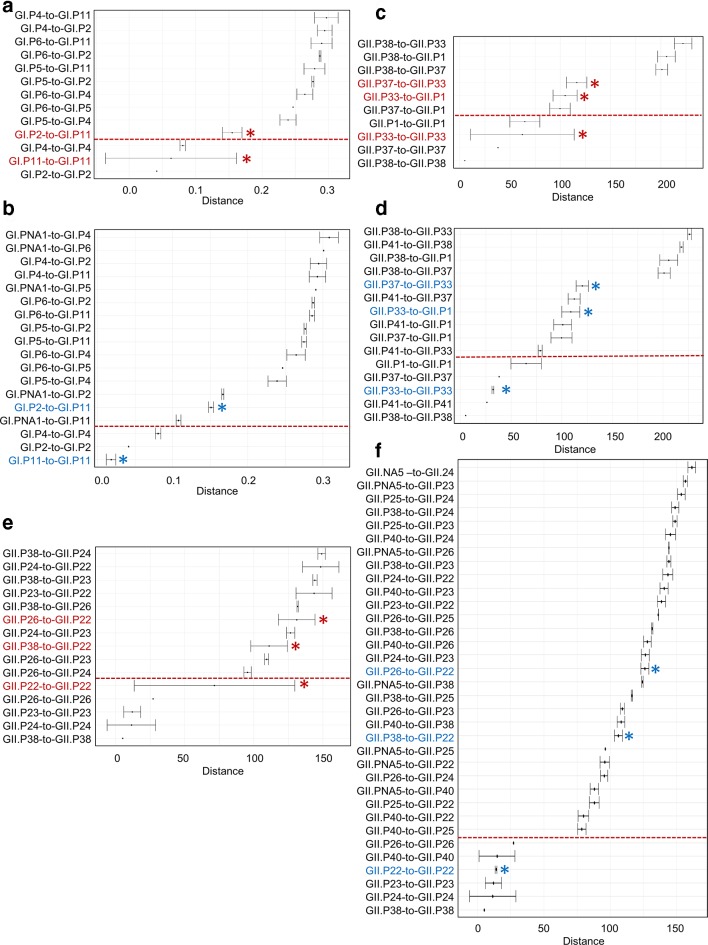
Fig. 4.
Patristic and P-distance comparison of GI.P11 (previously GI.Pb) (a and b), GII.P33 viruses (c and d) and GII.P22 viruses (e and f) with their phylogenetically closest P-types. Error bars represent 2xsd for each P-type comparison. The red * and font indicate the overlap of 2×sd error bars for viruses within a cluster and its closest P-types (a, b and e). After re-assigning GI.P11 viruses into two clusters (GI.P11 and GI.PNA1) (c), GII.P33 into GII.P33 and GII.P41 (d) and GII.P22 into three clusters (GII.P22, GII.P40 and GII.PNA5) (f), no overlap of 2×sd error bars within or in-between P-types was present (blue * and font). Y-axis represents comparison of distances within and in-between P-types. Below the dotted line, distances within P-type are indicated and above the dotted line, distances between P-types are indicated. (2×sd criteria: phylogenetic distances of sequences within a P-type should not overlap with distances between different P-types.)