Abstract
BACKGROUND
Spinal Muscular Atrophy (SMA) is a common autosomal recessive neuromuscular disease characterized by defects of lower motor neurons. More than 95% of SMA patients show homozygous deletion for the survival motor neuron 1 (SMN1) gene. For the screening of SMN1 deletion using dried blood spot (DBS), we developed a new combined system with real-time “modified competitive oligonucleotide priming”-polymerase chain reaction (mCOP-PCR) and PCR restriction fragment length polymorphism (PCR-RFLP). Although our real-time mCOP-PCR method is secured enough to be gene-specific, its amplification efficiency is not as good because the reverse primers carry a nucleotide mismatched with the sequence of the pre-amplified product. The mismatch has consequently been generated in the process of introducing a restriction enzyme site in the pre-amplified products for PCR-RFLP.
METHOD
DBS samples of the subjects were stored at room temperature for a period of less than one year. Each subject had already been genotyped by the first PCR-RFLP using fresh blood DNA. SMN1/SMN2 exon 7 was collectively amplified using conventional PCR (targeted pre-amplification). Pre-amplified products were used as template in the real-time mCOP-PCR, and, on the other hand, were digested with DraI enzyme (PCR-RFLP). To improve the amplification efficiency of mCOP-PCR, one nucleotide change was introduced in the original reverse primers (SMN1-COP and SMN2-COP) to eliminate the mismatched nucleotide.
RESULTS
The real-time mCOP-PCR with a new primer (SMN1-COP-DRA or SMN2-COP-DRA) more rapidly and specifically amplified SMN1 and SMN2, and clearly demonstrated SMN1 deletion in an SMA patient. With the new primers, the amplification efficiencies of real-time mCOP-PCR were improved and the Cq values of SMN1 (+) and SMN2 (+) samples were significantly lowered.
CONCLUSION
In the advanced version of our screening system for homozygous SMN1 deletion using DBS, the real-time mCOP-PCR with newly-designed reverse primers demonstrated the presence or absence of SMN1 and SMN2 within a shorter time, and the results were easily tested by PCR-RFLP. This rapid and accurate screening system will be useful for detection of newborn infants with SMA.
Keywords: spinal muscular atrophy, SMN1, SMN2, mCOP-PCR, targeted pre-amplification, PCR-RFLP
INTRODUCTION
Spinal Muscular Atrophy (SMA) is a common autosomal recessive neuromuscular disease characterized by defects of lower motor neurons. The incidence of the disease is 1 in 10,000 newborns, and its carrier frequency is 1 in 50 [9]. In 1995, the SMN genes SMN1 and SMN2 were identified as SMA-related genes in chromosome 5q [2, 7]. SMN1, which produces the SMN protein (SMN), is present in all healthy individuals. However, more than 95% of SMA patients show homozygous SMN1 deletion, while the remaining patients harbor some deleterious mutations in SMN1 [6]. SMN2, a gene highly homologous to SMN1, also produces small amount of SMN and modifies the SMA phenotype [3]. It is now believed that SMN1 is an SMA-causing gene and SMN2 an SMA-modifying gene, and that low levels of SMN protein cause SMA [3].
SMA has been recognized as an incurable disease. Although recent advances in respiratory care for neuromuscular disease have improved the survival period of SMA patients, there has yet been no treatment based on the pathogenesis of SMA. However, improved understanding of the molecular mechanisms of SMN2 expression has spurred the development of therapeutic compounds. In 2016, clinical trial results of intrathecal administration of an antisense-oligo, nusinersen, demonstrated encouraging clinical efficacy of the drug [4], leading to its approval by the United States Food and Drug Administration. We are now about to enter an era of the possibility of SMA treatment.
Thus, detection of infants with SMN1-deletion will become more important. By foreseeing future requirements, some researchers tried to establish a rapid method to differentiate between SMN1 and SMN2, which may be applied to newborn screening for SMA [11]. Screening policy varies from country to country, but with effective treatment to cure or arrest the progression of SMA, newborn screening for the disease may be warranted in every country. We also developed a new screening system with real-time “modified competitive oligonucleotide priming”-polymerase chain reaction (mCOP-PCR) using dried blood spot (DBS) [1, 10], and combined it with PCR-RFLP [12] for the purpose of confirming the results of real-time mCOP-PCR. PCR restriction fragment length polymorphism (PCR-RFLP) analysis is a time-consuming method, but it has been widely used and established as a method to differentiate SMN1 and SMN2.
Although the real-time mCOP-PCR in our combination system is secure enough to differentiate SMN1 and SMN2, its amplification efficiency is not as good due to the fact that the reverse primers carry a nucleotide mismatched with the sequence of the pre-amplified product [8]. The mismatch has consequently been generated in the process of introducing a restriction enzyme site in the pre-amplified products for PCR-RFLP. In this study, we developed a more rapid screening system for homozygous SMN1 deletion. Here, newly-designed reverse primers were used in the real-time mCOP-PCR in the combination system with PCR-RFLP, leading to improved amplification efficiency.
MATERIALS AND METHODS
DNA sample preparation
DNA samples were extracted from three individuals (two controls and one SMA patient) from a dried blood spot (DBS) on filter paper by the method of Kato et al. [5]. Each individual had been genotyped by PCR-RFLP using fresh blood DNA. Prior to analyses, informed consent was obtained from the patient’s families. The study was approved by the Ethics Committee of Kobe University Graduate School of Medicine.
Targeted pre-amplification
Targeted pre-amplification of the sequence containing SMN1/2 exon 7 was performed by conventional PCR using GeneAmp® PCR System 9700 (Applied Biosystems; Thermo Fisher Scientific, Foster City, CA, USA). Two μl of template solution (200~300 ng DNA from DBS) was directly added to PCR mixture (total volume, 28 μl) containing 1× PCR buffer [final concentration], 2 mM MgCl2 [final concentration], 0.1 mM of each dNTP, 0.3 μM of each primer (R111 and X7-Dra), and 1.0 U FastStart Taq DNA polymerase (Roche Applied Science, Mannheim, Germany). The primer sequences are shown in Table 1. The PCR conditions were: (1) initial denaturation at 94°C for 7 min; (2) 40 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min; (3) additional extension at 72°C for 7 min; and (4) hold at 10°C. Following this, an aliquot of pre-amplified product was electrophoresed on a 4% agarose gel in 1× TBE buffer, and visualized by Midori-Green Advance staining (Nippon Genetics, Tokyo, Japan).
Table I.
Sequences of forward and reverse primers used in this study
| Purpose | Forward | Reverse |
|---|---|---|
| Pre-amplification | R111: AGACTATCAACTTAATTTCTGATCA | SMN1/2: X7-Dra: CCTTCCTTCTTTTTGATTTTGTTT |
| mCOP-PCR | R111: AGACTATCAACTTAATTTCTGATCA |

|
| Advanced mCOP-PCR | R111: AGACTATCAACTTAATTTCTGATCA |

|
Sequences of forward and reverse primers used in this study. For pre-amplification and PCR-RFLP. R111 and X7-Dra were used. For the regular version of mCOP-PCR. R111 was used in combination with either SMN1-COP or SMN2-COP. For the advanced version of mCOP-PCR R111 was used in combination with either SMN1-COP-DRA or SMN2-COP-DRA. Gene-specific nucleotides are in red, and a DraI site is underlined.
Real-time mCOP-PCR
The real-time mCOP-PCR was performed using the Light Cycler® 96 Real-time PCR system (Roche Molecular Systems, Inc). After 100-fold dilution of pre-amplified product, 4 μl diluted solution was added to the PCR mixture (final volume, 30 μl) containing 1 × PCR buffer (final concentration), 2 mM MgCl2 [final concentration], 0.1 mM of each dNTP, 0.3 μM of common forward primer (R111), 0.3 μM of gene-specific reverse primer (SMN1-COP / SMN2-COP, SMN1-COP-DRA / SMN2-COP-DRA), 1.0 U FastStart Taq DNA polymerase and 1.5 μl of 20× EvaGreen® Dye (Biotium, Hayward, CA, USA). The primer sequences are shown in Table 1. The PCR conditions were: (1) initial denaturation at 94°C for 7 min; (2) 30 or 40 cycles of denaturation at 94°C for 1 min, annealing at 37°C for 1 min, and extension at 72°C for 1 min; (3) additional extension at 72°C for 7 min; and (4) hold at 10°C. The cycle number was 20 for SMN1-COP-DRA, 20 for SMN2-COP-DRA, 30 for SMN1-COP, and 40 for or SMN2-COP.
PCR-RFLP
DraI site was introduced into SMN2 product during the targeted pre-amplification step. The pre-amplified SMN2 product was digested by overnight incubation with DraI. More specifically, 8 μl of pre-amplified products was added to the enzyme solution (final volume, 20 μl) containing 1 × buffer M [final concentration] and 1 μl of DraI (15 U/μl) (Takara Biomedicals, Shiga, Japan), and the mixture was incubated at 37°C overnight. Then, an aliquot of digested pre-amplified product was electrophoresed on a 4% agarose gel in 1×TBE buffer, and visualized by Midori-Green Advance staining (Nippon Genetics). After DraI digestion, the pre-amplified SMN2 product (187 bp) generated two fragments of 163 bp and 24 bp, while the pre-amplified SMN1 product (187 bp) did not undergo DraI digestion and remained the same size as the non-digested one.
RESULTS
Amplification efficiency of real-time mCOP-PCR
The original primers, SMN1-COP and SMN2-COP, carried a nucleotide mismatched with the pre-amplified product, while the newly-designed primers, SMN1-COP-DRA and SMN2-COP-DRA, did not. Here, we used a control DBS-DNA sample with a genotype of [SMN1 (+) / SMN2 (+)]. To evaluate the effect of reverse primers on amplification, we applied 4, 6, 8 and 10 μl of 100-fold diluted “pre-amplified product” solution to real-time mCOP-PCR with each reverse primer, and determined the amplification efficiency.
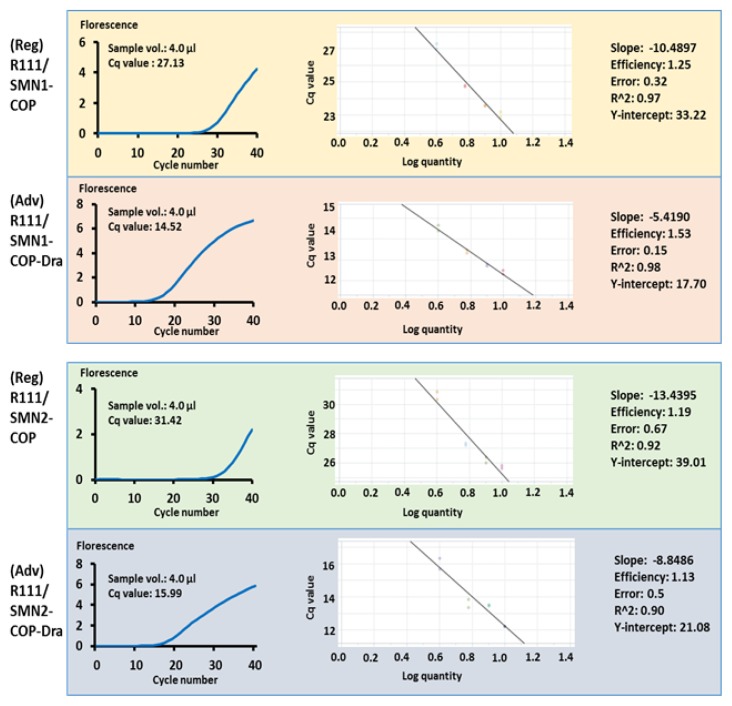
As shown in Figure 1, amplification efficiency of mCOP-PCR with SMN1-COP-DRA was better than that with SMN1-COP. The quantification cycle (Cq) values of 4 μl of the 100-fold diluted “pre-amplified product” solution with SMN1-COP-DRA were 14.52, while the Cq values of 4 μl of the solution with SMN1-COP were 27.13.
Figure 1.
Cq values (4 μl of 100-fold diluted “pre-amplified product” solution was used in this study), “Cq-Log quantity” curves and amplification efficiencies of two versions of real-time mCOP-PCR. Regular version (Reg) of real-time mCOP-PCR was performed with R111/SMN1-COP or R111/SMN2-COP, and advanced version (Adv) of real-time mCOP-PCR was performed with R111/SMN1-COP-DRA or R111/SMN2-COP-DRA.
The same is true of SMN2-COP-DRA; the amplification efficiency was better than that with SMN2-COP (Figure 1). The Cq values of 4 μl of the solution with SMN2-COP-DRA were 15.99, while the Cq values of 4 μl of the solution with SMN2-COP were 31.42.
Genotyping analysis using real-time mCOP-PCR
Here, we performed genotyping analysis using real-time mCOP-PCR with SMN1-COP-DRA and SMN2-COP-DRA. All three of the samples had been genotyped by PCR-RFLP using fresh blood DNA.
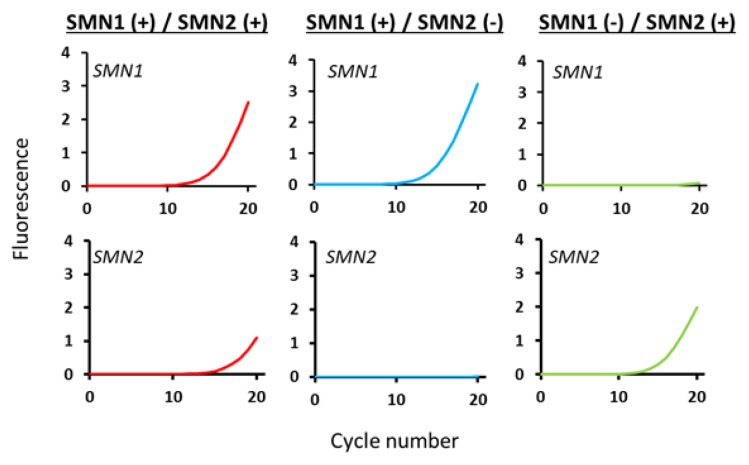
For SMN1 amplification in a real-time mCOP-PCR with SMN1-COP-DRA, the Cq values for SMN1 positive samples [SMN1 (+) / SMN2 (+) and SMN1 (+) / SMN2 (−)] were less than 15 (Figure 2). However, the Cq value for SMN1 negative sample [SMN1 (−) / SMN2 (+)] could not be obtained, as no apparent elevation of amplification curve was observed prior to 30 cycles. Thus, the presence or absence of SMN1 was unambiguously detected in spite of the presence of SMN2.
Figure 2.
Genotyping by real-time mCOP-PCR with R111/SMN1-COP-DRA and R111/SMN2-COP-DRA. Real-time mCOP-PCR amplification of the samples with [SMN1 (+) / SMN2 (+)], [SMN1 (+) / SMN2 (−)] and [SMN1 (−) / SMN2 (+)]. The samples with [SMN1 (+) / SMN2 (+)], [SMN1 (+) / SMN2 (−)] showed amplification with R111/SMN1-COP-DRA, while the samples with [SMN1 (−) / SMN2 (+)] showed no amplification with the primer set. The samples with [SMN1 (+) / SMN2 (+)], [SMN1 (−) / SMN2 (+)] showed amplification with R111/SMN2-COP-DRA, while the samples with [SMN1 (+) / SMN2 (−)] showed no amplification with the primer set.
For SMN2 amplification in a real-time mCOP-PCR with SMN2-COP-DRA, the Cq values for SMN2 positive samples [SMN1 (+) / SMN2 (+) and SMN1 (−) / SMN2 (+)] were less than 17 (Figure 2). However, the Cq value for SMN2 negative sample [SMN1 (+) / SMN2 (−)] could not be obtained, as no apparent elevation of amplification curve was observed prior to 35 cycles. Thus, the presence or absence of SMN2 was unambiguously detected in spite of the presence of SMN1.
To confirm the results of real-time mCOP-PCR, we performed the second PCR-RFLP assay using the pre-amplified product (the first PCR-RFLP assay was performed using fresh blood DNA, as described in the Methods section). The results of real-time mCOP-PCR were completely consistent with the results of the first and second PCR-RFLP assay (data not shown).
The results of the other cases examined in this study, including healthy controls and SMA patients, were completely consistent with the results shown in the figures (data not shown).
DISCUSSION
PCR-RFLP may resolve the ambiguous results of real-time mCOP-PCR
Our screening system for homozygous SMN1 deletion consists of real-time mCOP-PCR and PCR-RFLP. Real-time mCOP-PCR is a very rapid, accurate method to distinguish SMN1 and SMN2. However, it is necessary to ensure that the test samples are preserved from contamination from other samples’ DNA. Nevertheless, it should be considered that low-level contamination may hamper the correct diagnosis, because our real-time mCOP-PCR method is so sensitive that occasionally, false positive results can be obtained, leading to mis-classification of an SMN1-deleted case as an SMN1-retained one.
On the other hand, PCR-RFLP, which is usually followed by gel-electrophoresis, is time-consuming and less sensitive compared to real-time mCOP-PCR, but low-level contamination may not affect the diagnosis. Thus, PCR-PFLP may be necessary to check the ambiguous results of real-time mCOP-PCR. Combination of real-time mCOP-PCR and PCR-RFLP may improve the accuracy of the disease screening.
Newly-designed primers improve amplification efficiency
Pre-amplification with a primer set for PCR-RFLP, R111 and X7-Dra, enabled us to check the results of real-time mCOP-PCR easily, because direct digestion of the pre-amplification product completes PCR-RFLP. X7-Dra was a mismatched primer which introduced a DraI site into the sequence of SMN2 exon 7.
Thus, the pre-amplified product contained a nucleotide mismatched with the genuine sequence of SMN1 and SMN2. Consequently, the gene-specific mCOP-PCR primers, SMN1-COP and SMN2-COP, contained an additional nucleotide mismatched with the sequence of the pre-amplified product.
In this study, we decided to eliminate a nucleotide mismatched (C) to the sequence of the pre-amplified product from the original primers, SMN1-COP and SMN2-COP, and designed new primers, SMN1-COP-DRA and SMN2-COP-DRA. Elimination of the mismatched nucleotide improved the amplification efficiency (Figure 1), but it did not affect the gene-specific amplification. This advanced version of our screening system for homozygous SMN1 deletion is much superior to the current version of our system.
Conclusion
In the advanced version of our screening system for homozygous SMN1 deletion using DBS, the real-time mCOP-PCR with newly-designed reverse primers demonstrated the presence or absence of SMN1 and SMN2 in the earlier cycles of amplification, and the results were easily tested by PCR-RFLP. This rapid and accurate screening system will be useful for detection of SMA in newborn infants.
ACKNOWLEDGMENTS
This research was supported in part by the Practical Research Project for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development, Grant No. 16ek0109086h0002 (title “Practical study for multicenter cooperative and investigator initiated clinical trial using valproic acid in childhood onset spinal muscular atrophy”) and by the Ministry of Education, Culture, Sports, Science and Technology, Japan, Grant No. 25461549.
REFERENCES
- 1.Ar Rochmah M, Harahap NIF, Niba ETE, et al. Genetic screening of spinal muscular atrophy using a real-time modified COP-PCR technique with dried blood-spot DNA. Brain Dev. 2017;39:774–782. doi: 10.1016/j.braindev.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 2.Brzustowicz LM, Lehner T, Castilla LH, et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2–13.3. Nature. 1990;344:540–541. doi: 10.1038/344540a0. [DOI] [PubMed] [Google Scholar]
- 3.Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finkel RS, Chiriboga CA, Vajsar J, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388:3017–3026. doi: 10.1016/S0140-6736(16)31408-8. [DOI] [PubMed] [Google Scholar]
- 5.Kato N, Sa’Adah N, Ar Rochmah M, et al. SMA screening system using dried blood spots on filter paper: application of COP-PCR to the SMN1 deletion test. Kobe J Med Sci. 2014;60:E78–85. [PubMed] [Google Scholar]
- 6.Lefebvre S, Bürglen L, Reboullet S, et al. Identification and characterization of a Spinal Muscular Atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 7.Melki J, Abdelhak S, Sheth P, et al. Gene for chronic proximal spinal muscular atrophies maps to chromosome 5q. Nature. 1990;344:767–768. doi: 10.1038/344767a0. [DOI] [PubMed] [Google Scholar]
- 8.Niba ETE, Ar Rochmah M, Harahap NIF, et al. Spinal Muscular Atrophy: New Screening System with Real-Time mCOP-PCR and PCR-RFLP for SMN1 Deletion. Kobe J Med Sci. 2019;65:E44–48. [PMC free article] [PubMed] [Google Scholar]
- 9.Nurputra DK, Lai PS, Harahap NI, et al. Spinal Muscular Atrophy: From Gene Discovery to Clinical Trials. Ann Hum Genet. 2013;77:435–463. doi: 10.1111/ahg.12031. [DOI] [PubMed] [Google Scholar]
- 10.Shinohara M, Ar Rochmah M, Nakanishi K, et al. New, Improved Version of the mCOP-PCR Screening System for Detection of Spinal Muscular Atrophy Gene (SMN1) Deletion. Kobe J Med Sci. 2017;63:E37–40. [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor JL, Lee FK, Yazdanpanah GK, et al. Newborn Blood Spot Screening Test Using Multiplexed Real-Time PCR to Simultaneously Screen for Spinal Muscular Atrophy and Severe Combined Immunodeficiency. Clin Chem. 2015;61:412–419. doi: 10.1373/clinchem.2014.231019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Steege G, Grootscholten PM, van der Vlies P, et al. PCR-based DNA test to confirm clinical diagnosis of autosomal recessive spinal muscular atrophy. The Lancet. 1995;345:985–986. [PubMed] [Google Scholar]