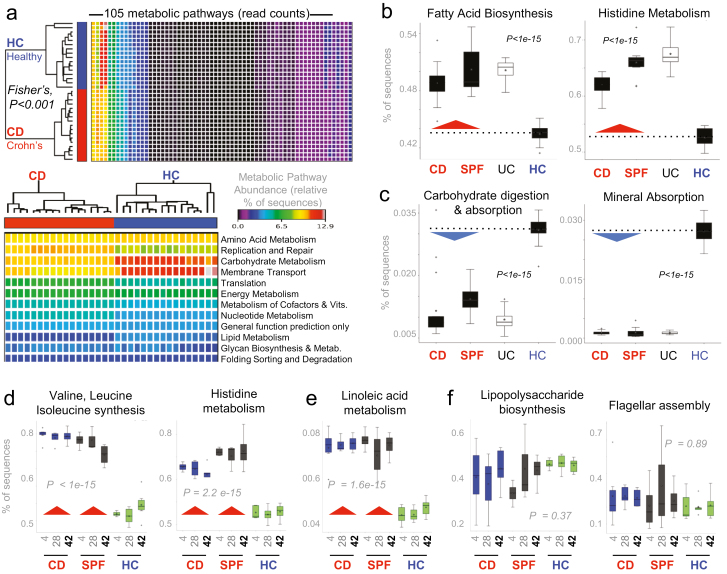
FIGURE 7.
Metabolic microbiome predictive analysis reveals chronically abnormal pathways in FMT SAMP mice. Statistical analyses of metagenomic profiles (STAMP)28 of relative abundance values for various KEGG metabolic pathways encoded in mice fecal samples (Exp. A). A, Heatmap illustrates that metabolic predictions are reproducible across samples and that the CD and HC group cluster together with apparent differences for some pathways. B, Boxplots of sequence abundance (ANOVA with multiple tests correction by Benjamin-Hochberg FDR) for representative KEGG pathways deemed relevant for driving severity of inflammation in experimental CD. Boxplots show that some paths are enriched while others are deficient in the microbiota, irrespective of whether it originates from a human with CD or a mouse with severe CD-ileitis. C–E, Boxplots of sequence abundance illustrates the consistent, inverse differences in microbiota functionality for CD donor mice (severely inflamed mice; CD donor A#1, see Fig. 1C) and SPF donor mice to mice that received feces from the healthy non-IBD donor (HC donor A#7; see Fig. 1C) over time (days 4, 28, and 42). Plots illustrate that BCAAs (valine, leucine, isoleucine), histidine, and linoleic acid remain chronically synthesized and degraded (Supplementary Fig. 14) through the study period, but that there is no difference in virulence markers (LPS, flagellin; panel E) between groups (post hoc study power calculations achieved >0.80).45 Chronically abnormal metabolic pathways in the gut microbiota could be chronic drivers or biomarkers that precede CD-like ileitis.